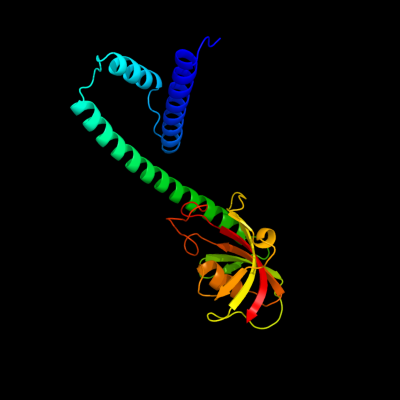
| 1 | d1q6ha_
|
|
 |
100.0 |
99 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
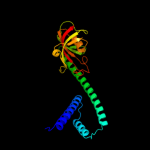
| 2 | c1q6uA_
|
|
 |
100.0 |
100 |
PDB header:isomerase
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase fkpa;
PDBTitle: crystal structure of fkpa from escherichia coli
|
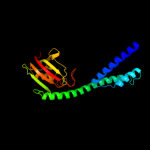
| 3 | d1fd9a_
|
|
 |
100.0 |
37 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 4 | c2vcdA_
|
|
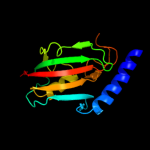 |
100.0 |
41 |
PDB header:isomerase
Chain: A: PDB Molecule:outer membrane protein mip;
PDBTitle: solution structure of the fkbp-domain of legionella2 pneumophila mip in complex with rapamycin
|
| 5 | d1jvwa_
|
|
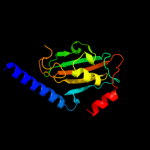 |
100.0 |
38 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 6 | c3oe2A_
|
|
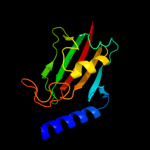 |
100.0 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl-prolyl cis-trans isomerase;
PDBTitle: 1.6 a crystal structure of peptidyl-prolyl cis-trans isomerase ppiase2 from pseudomonas syringae pv. tomato str. dc3000 (pspto dc3000)
|
| 7 | c2lgoA_
|
|
 |
100.0 |
45 |
PDB header:isomerase
Chain: A: PDB Molecule:fkbp;
PDBTitle: solution nmr structure of a fkbp-type peptidyl-prolyl cis-trans2 isomerase from giardia lamblia, seattle structural genomics center3 for infectious disease target gilaa.00840.a
|
| 8 | d1yata_
|
|
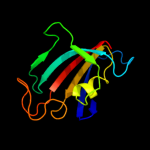 |
100.0 |
46 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 9 | c2ke0A_
|
|
 |
100.0 |
50 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl-prolyl cis-trans isomerase;
PDBTitle: solution structure of peptidyl-prolyl cis-trans isomerase from2 burkholderia pseudomallei
|
| 10 | d1q1ca1
|
|
 |
100.0 |
42 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 11 | c2jwxA_
|
|
 |
100.0 |
20 |
PDB header:apoptosis, isomerase
Chain: A: PDB Molecule:fk506-binding protein 8 variant;
PDBTitle: solution structure of the n-terminal domain of human fkbp382 (fkbp38ntd)
|
| 12 | d1c9ha_
|
|
 |
100.0 |
46 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 13 | d1r9ha_
|
|
 |
100.0 |
39 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 14 | c2f4eB_
|
|
 |
100.0 |
23 |
PDB header:signaling protein
Chain: B: PDB Molecule:atfkbp42;
PDBTitle: n-terminal domain of fkbp42 from arabidopsis thaliana
|
| 15 | c3o5fA_
|
|
 |
100.0 |
47 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl-prolyl cis-trans isomerase fkbp5;
PDBTitle: fk1 domain of fkbp51, crystal form vii
|
| 16 | c2vn1A_
|
|
 |
100.0 |
35 |
PDB header:isomerase
Chain: A: PDB Molecule:70 kda peptidylprolyl isomerase;
PDBTitle: crystal structure of the fk506-binding domain of plasmodium2 falciparum fkbp35 in complex with fk506
|
| 17 | c1rouA_
|
|
 |
100.0 |
43 |
PDB header:rotamase (isomerase)
Chain: A: PDB Molecule:fkbp59-i;
PDBTitle: structure of fkbp59-i, the n-terminal domain of a 59 kda2 fk506-binding protein, nmr, 22 structures
|
| 18 | d2ppna1
|
|
 |
100.0 |
49 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 19 | d1u79a_
|
|
 |
100.0 |
45 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 20 | d1kt0a3
|
|
 |
100.0 |
24 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 21 | d1kt1a3 |
|
not modelled |
100.0 |
23 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 22 | c3b7xA_ |
|
not modelled |
100.0 |
29 |
PDB header:isomerase
Chain: A: PDB Molecule:fk506-binding protein 6;
PDBTitle: crystal structure of human fk506-binding protein 6
|
| 23 | c3o5dB_ |
|
not modelled |
100.0 |
46 |
PDB header:isomerase
Chain: B: PDB Molecule:peptidyl-prolyl cis-trans isomerase fkbp5;
PDBTitle: crystal structure of a fragment of fkbp51 comprising the fk1 and fk22 domains
|
| 24 | c1q1cA_ |
|
not modelled |
99.9 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:fk506-binding protein 4;
PDBTitle: crystal structure of n(1-260) of human fkbp52
|
| 25 | c2d9fA_ |
|
not modelled |
99.9 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:fk506-binding protein 8 variant;
PDBTitle: solution structure of ruh-047, an fkbp domain from human2 cdna
|
| 26 | d1pbka_ |
|
not modelled |
99.9 |
47 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 27 | c2pbcD_ |
|
not modelled |
99.9 |
46 |
PDB header:isomerase
Chain: D: PDB Molecule:fk506-binding protein 2;
PDBTitle: fk506-binding protein 2
|
| 28 | d1q1ca2 |
|
not modelled |
99.9 |
24 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 29 | c3jxvA_ |
|
not modelled |
99.9 |
31 |
PDB header:isomerase
Chain: A: PDB Molecule:70 kda peptidyl-prolyl isomerase;
PDBTitle: crystal structure of the 3 fkbp domains of wheat fkbp73
|
| 30 | c2if4A_ |
|
not modelled |
99.9 |
20 |
PDB header:signaling protein
Chain: A: PDB Molecule:atfkbp42;
PDBTitle: crystal structure of a multi-domain immunophilin from2 arabidopsis thaliana
|
| 31 | d1kt0a2 |
|
not modelled |
99.9 |
44 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 32 | d1kt1a2 |
|
not modelled |
99.9 |
41 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 33 | c1qz2B_ |
|
not modelled |
99.9 |
24 |
PDB header:isomerase/chaperone
Chain: B: PDB Molecule:fk506-binding protein 4;
PDBTitle: crystal structure of fkbp52 c-terminal domain complex with2 the c-terminal peptide meevd of hsp90
|
| 34 | c1kt0A_ |
|
not modelled |
99.9 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:51 kda fk506-binding protein;
PDBTitle: structure of the large fkbp-like protein, fkbp51, involved in steroid2 receptor complexes
|
| 35 | c2kr7A_ |
|
not modelled |
99.8 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase slyd;
PDBTitle: solution structure of helicobacter pylori slyd
|
| 36 | c2k8iA_ |
|
not modelled |
99.7 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl-prolyl cis-trans isomerase;
PDBTitle: solution structure of e.coli slyd
|
| 37 | c2kfwA_ |
|
not modelled |
99.7 |
27 |
PDB header:isomerase
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase
PDBTitle: solution structure of full-length slyd from e.coli
|
| 38 | c3pr9A_ |
|
not modelled |
99.7 |
31 |
PDB header:chaperone
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase;
PDBTitle: structural analysis of protein folding by the methanococcus jannaschii2 chaperone fkbp26
|
| 39 | d1ix5a_ |
|
not modelled |
99.7 |
36 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 40 | c3prdA_ |
|
not modelled |
99.7 |
36 |
PDB header:chaperone, isomerase
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase;
PDBTitle: structural analysis of protein folding by the methanococcus jannaschii2 chaperone fkbp26
|
| 41 | c3cgnA_ |
|
not modelled |
99.6 |
35 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl-prolyl cis-trans isomerase;
PDBTitle: crystal structure of thermophilic slyd
|
| 42 | c1hxvA_ |
|
not modelled |
99.5 |
20 |
PDB header:chaperone
Chain: A: PDB Molecule:trigger factor;
PDBTitle: ppiase domain of the mycoplasma genitalium trigger factor
|
| 43 | d1hxva_ |
|
not modelled |
99.5 |
20 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 44 | d1l1pa_ |
|
not modelled |
99.4 |
23 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 45 | d1w26a3 |
|
not modelled |
99.3 |
22 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 46 | d1t11a3 |
|
not modelled |
99.3 |
21 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 47 | c1w26B_ |
|
not modelled |
98.6 |
20 |
PDB header:chaperone
Chain: B: PDB Molecule:trigger factor;
PDBTitle: trigger factor in complex with the ribosome forms a2 molecular cradle for nascent proteins
|
| 48 | c1t11A_ |
|
not modelled |
98.4 |
20 |
PDB header:chaperone
Chain: A: PDB Molecule:trigger factor;
PDBTitle: trigger factor
|
| 49 | c3gtyX_ |
|
not modelled |
97.6 |
19 |
PDB header:chaperone/ribosomal protein
Chain: X: PDB Molecule:trigger factor;
PDBTitle: promiscuous substrate recognition in folding and assembly activities2 of the trigger factor chaperone
|
| 50 | c3htxA_ |
|
not modelled |
96.4 |
13 |
PDB header:transferase/rna
Chain: A: PDB Molecule:hen1;
PDBTitle: crystal structure of small rna methyltransferase hen1
|
| 51 | d1jnsa_ |
|
not modelled |
46.7 |
12 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 52 | c2a2bA_ |
|
not modelled |
41.4 |
40 |
PDB header:antibiotic
Chain: A: PDB Molecule:bacteriocin curvacin a;
PDBTitle: curvacin a
|
| 53 | d2diga1 |
|
not modelled |
29.8 |
43 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:Tudor domain |
| 54 | d1pina2 |
|
not modelled |
29.2 |
10 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 55 | d2pv2a1 |
|
not modelled |
28.8 |
17 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 56 | c2rqsA_ |
|
not modelled |
28.8 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:parvulin-like peptidyl-prolyl isomerase;
PDBTitle: 3d structure of pin from the psychrophilic archeon cenarcheaum2 symbiosum (cspin)
|
| 57 | d1j6ya_ |
|
not modelled |
26.9 |
14 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 58 | d1ueba1 |
|
not modelled |
26.8 |
23 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 59 | c3gpkA_ |
|
not modelled |
23.3 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:ppic-type peptidyl-prolyl cis-trans isomerase;
PDBTitle: crystal structure of ppic-type peptidyl-prolyl cis-trans isomerase2 domain at 1.55a resolution.
|
| 60 | c3a5zF_ |
|
not modelled |
21.8 |
17 |
PDB header:ligase
Chain: F: PDB Molecule:elongation factor p;
PDBTitle: crystal structure of escherichia coli genx in complex with elongation2 factor p
|
| 61 | d1o65a_ |
|
not modelled |
20.3 |
23 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:MOSC (MOCO sulphurase C-terminal) domain |
| 62 | c2eqmA_ |
|
not modelled |
20.1 |
9 |
PDB header:transcription
Chain: A: PDB Molecule:phd finger protein 20-like 1;
PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1 [homo sapiens]
|
| 63 | c1yw5A_ |
|
not modelled |
18.0 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl prolyl cis/trans isomerase;
PDBTitle: peptidyl-prolyl isomerase ess1 from candida albicans
|
| 64 | d1m5ya3 |
|
not modelled |
17.8 |
14 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 65 | d1gpla1 |
|
not modelled |
16.6 |
14 |
Fold:Lipase/lipooxygenase domain (PLAT/LH2 domain)
Superfamily:Lipase/lipooxygenase domain (PLAT/LH2 domain)
Family:Colipase-binding domain |
| 66 | d1orua_ |
|
not modelled |
16.0 |
36 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:MOSC (MOCO sulphurase C-terminal) domain |
| 67 | d1fcda2 |
|
not modelled |
15.1 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 68 | c3qgaD_ |
|
not modelled |
14.8 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:fusion of urease beta and gamma subunits;
PDBTitle: 3.0 a model of iron containing urease urea2b2 from helicobacter2 mustelae
|
| 69 | d2hd9a1 |
|
not modelled |
14.7 |
22 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:Atu2648/PH1033-like |
| 70 | c1zk6A_ |
|
not modelled |
12.6 |
9 |
PDB header:isomerase
Chain: A: PDB Molecule:foldase protein prsa;
PDBTitle: nmr solution structure of b. subtilis prsa ppiase
|
| 71 | d1ejxb_ |
|
not modelled |
11.1 |
32 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 72 | c3rfwA_ |
|
not modelled |
11.0 |
16 |
PDB header:chaperone
Chain: A: PDB Molecule:cell-binding factor 2;
PDBTitle: the virulence factor peb4 and the periplasmic protein cj1289 are two2 structurally-related sura-like chaperones in the human pathogen3 campylobacter jejuni
|
| 73 | d2f8la1 |
|
not modelled |
10.6 |
18 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:N-6 DNA Methylase-like |
| 74 | d2vj0a1 |
|
not modelled |
10.6 |
17 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Clathrin adaptor appendage domain
Family:Alpha-adaptin ear subdomain-like |
| 75 | d2ih2a1 |
|
not modelled |
9.8 |
18 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:DNA methylase TaqI, N-terminal domain |
| 76 | d4ubpb_ |
|
not modelled |
9.8 |
28 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 77 | d1e9ya1 |
|
not modelled |
9.3 |
36 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 78 | c3q1jA_ |
|
not modelled |
8.6 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:phd finger protein 20;
PDBTitle: crystal structure of tudor domain 1 of human phd finger protein 20
|
| 79 | c2jzvA_ |
|
not modelled |
7.5 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:foldase protein prsa;
PDBTitle: solution structure of s. aureus prsa-ppiase
|
| 80 | d1etha1 |
|
not modelled |
7.3 |
14 |
Fold:Lipase/lipooxygenase domain (PLAT/LH2 domain)
Superfamily:Lipase/lipooxygenase domain (PLAT/LH2 domain)
Family:Colipase-binding domain |
| 81 | c1f8aB_ |
|
not modelled |
7.2 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:peptidyl-prolyl cis-trans isomerase nima-
PDBTitle: structural basis for the phosphoserine-proline recognition2 by group iv ww domains
|
| 82 | d1qfja1 |
|
not modelled |
7.1 |
10 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 83 | c3on1A_ |
|
not modelled |
6.9 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:bh2414 protein;
PDBTitle: the structure of a protein with unknown function from bacillus2 halodurans c
|
| 84 | d1eq3a_ |
|
not modelled |
6.9 |
13 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 85 | c1mv3A_ |
|
not modelled |
6.6 |
28 |
PDB header:endocytosis/exocytosis
Chain: A: PDB Molecule:myc box dependent interacting protein 1;
PDBTitle: nmr structure of the tumor suppressor bin1: alternative2 splicing in melanoma and interaction with c-myc
|
| 86 | c3ikmD_ |
|
not modelled |
6.6 |
50 |
PDB header:transferase
Chain: D: PDB Molecule:dna polymerase subunit gamma-1;
PDBTitle: crystal structure of human mitochondrial dna polymerase2 holoenzyme
|
| 87 | d2j44a1 |
|
not modelled |
6.6 |
50 |
Fold:Prealbumin-like
Superfamily:Starch-binding domain-like
Family:PUD-like |
| 88 | c3oyyA_ |
|
not modelled |
6.4 |
14 |
PDB header:translation
Chain: A: PDB Molecule:elongation factor p;
PDBTitle: structure of pseudomonas aeruginosa elongation factor p
|
| 89 | c2p5dA_ |
|
not modelled |
6.3 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0310 protein mjecl36;
PDBTitle: crystal structure of mjecl36 from methanocaldococcus2 jannaschii dsm 2661
|
| 90 | c2wdtA_ |
|
not modelled |
6.2 |
20 |
PDB header:hydrolase/protein binding
Chain: A: PDB Molecule:ubiquitin carboxyl-terminal hydrolase l3;
PDBTitle: crystal structure of plasmodium falciparum uchl3 in complex2 with the suicide inhibitor ubvme
|
| 91 | d1js8a2 |
|
not modelled |
6.2 |
27 |
Fold:C-terminal domain of mollusc hemocyanin
Superfamily:C-terminal domain of mollusc hemocyanin
Family:C-terminal domain of mollusc hemocyanin |
| 92 | d2qqra2 |
|
not modelled |
6.2 |
18 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:Tudor domain |
| 93 | d1p7ka1 |
|
not modelled |
6.1 |
10 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 94 | d1xd3a_ |
|
not modelled |
6.0 |
20 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Ubiquitin carboxyl-terminal hydrolase UCH-L |
| 95 | d1v29a_ |
|
not modelled |
5.9 |
17 |
Fold:Nitrile hydratase alpha chain
Superfamily:Nitrile hydratase alpha chain
Family:Nitrile hydratase alpha chain |
| 96 | d2qqsa2 |
|
not modelled |
5.8 |
18 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:Tudor domain |
| 97 | c1e9zA_ |
|
not modelled |
5.8 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:urease subunit alpha;
PDBTitle: crystal structure of helicobacter pylori urease
|
| 98 | c1m5yB_ |
|
not modelled |
5.7 |
17 |
PDB header:isomerase, cell cycle
Chain: B: PDB Molecule:survival protein sura;
PDBTitle: crystallographic structure of sura, a molecular chaperone2 that facilitates outer membrane porin folding
|
| 99 | c3n42F_ |
|
not modelled |
5.5 |
29 |
PDB header:viral protein
Chain: F: PDB Molecule:e1 envelope glycoprotein;
PDBTitle: crystal structures of the mature envelope glycoprotein complex (furin2 cleavage) of chikungunya virus.
|