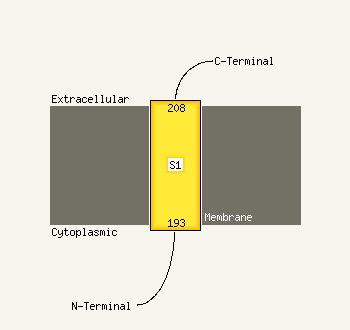
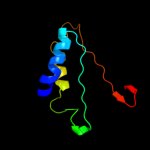
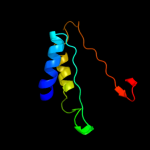
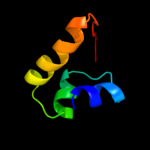
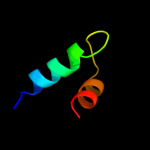
1 d1iuqa_
97.5
16
Fold: Glycerol-3-phosphate (1)-acyltransferaseSuperfamily: Glycerol-3-phosphate (1)-acyltransferaseFamily: Glycerol-3-phosphate (1)-acyltransferase2 d2a07f1
31.6
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain3 c3hqdB_
29.7
20
PDB header: motor proteinChain: B: PDB Molecule: kinesin-like protein kif11;PDBTitle: human kinesin eg5 motor domain in complex with amppnp and2 mg2+
4 c2jlhA_
28.3
18
PDB header: protein transportChain: A: PDB Molecule: yop proteins translocation protein u;PDBTitle: crystal structure of the cytoplasmic domain of yersinia2 pestis yscu n263a mutant
5 d2dpwa1
28.0
25
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: TTHA0179-like6 c3tevA_
26.7
22
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hyrolase, family 3;PDBTitle: the crystal structure of glycosyl hydrolase from deinococcus2 radiodurans r1
7 d1tr9a_
25.8
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: NagZ-like8 d2p6ra1
25.3
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RecQ helicase DNA-binding domain-like9 d1t3la2
21.7
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases10 c1e17A_
20.8
19
PDB header: dna binding domainChain: A: PDB Molecule: afx;PDBTitle: solution structure of the dna binding domain of the human2 forkhead transcription factor afx (foxo4)
11 c3s2wB_
19.7
18
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: the crystal structure of a marr transcriptional regulator from2 methanosarcina mazei go1
12 d1vyva2
18.7
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases13 d1t0hb_
18.3
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases14 d1vyua2
17.4
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases15 d1ry6a_
16.9
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins16 c1ry6A_
16.9
14
PDB header: transport proteinChain: A: PDB Molecule: internal kinesin;PDBTitle: crystal structure of internal kinesin motor domain
17 d1wj5a_
16.3
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: An Obfc1 domain18 d1z6ra1
16.1
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ROK associated domain19 d2c6ya1
15.9
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain20 d1mvoa_
15.5
14
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related21 c1sfeA_
not modelled
15.2
57
PDB header: dna-binding proteinChain: A: PDB Molecule: ada o6-methylguanine-dna methyltransferase;PDBTitle: ada o6-methylguanine-dna methyltransferase from escherichia coli
22 c2jk1A_
not modelled
14.7
7
PDB header: dna-bindingChain: A: PDB Molecule: hydrogenase transcriptional regulatory protein hupr1;PDBTitle: crystal structure of the wild-type hupr receiver domain
23 c3t6aB_
not modelled
14.5
15
PDB header: signaling proteinChain: B: PDB Molecule: breast cancer anti-estrogen resistance protein 3;PDBTitle: structure of the c-terminal domain of bcar3
24 c3c00B_
not modelled
14.4
11
PDB header: membrane protein, protein transportChain: B: PDB Molecule: escu;PDBTitle: crystal structural of the mutated g247t escu/spas c-terminal domain
25 c2k29A_
not modelled
14.1
13
PDB header: transcriptionChain: A: PDB Molecule: antitoxin relb;PDBTitle: structure of the dbd domain of e. coli antitoxin relb
26 c2wbeC_
not modelled
14.0
17
PDB header: structural proteinChain: C: PDB Molecule: bipolar kinesin krp-130;PDBTitle: kinesin-5-tubulin complex with amppnp
27 d1x88a1
not modelled
13.8
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins28 c3lk6A_
not modelled
13.2
25
PDB header: hydrolaseChain: A: PDB Molecule: lipoprotein ybbd;PDBTitle: beta-n-hexosaminidase n318d mutant (ybbd_n318d) from bacillus subtilis
29 d2qzga1
not modelled
13.0
12
Fold: Bromodomain-likeSuperfamily: Ta0600-likeFamily: Ta0600-like30 c1wrjA_
not modelled
12.9
57
PDB header: transferaseChain: A: PDB Molecule: methylated-dna--protein-cysteinePDBTitle: crystal structure of o6-methylguanine methyltransferase2 from sulfolobus tokodaii
31 c3g73A_
not modelled
12.5
11
PDB header: transcription/dnaChain: A: PDB Molecule: forkhead box protein m1;PDBTitle: structure of the foxm1 dna binding
32 c3sqlB_
not modelled
12.2
17
PDB header: hydrolaseChain: B: PDB Molecule: glycosyl hydrolase family 3;PDBTitle: crystal structure of glycoside hydrolase from synechococcus
33 d1q42a_
not modelled
12.2
39
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: NTF2-like34 c2vt1B_
not modelled
12.1
17
PDB header: membrane proteinChain: B: PDB Molecule: surface presentation of antigens protein spas;PDBTitle: crystal structure of the cytoplasmic domain of spa40, the2 specificity switch for the shigella flexneri type iii3 secretion system
35 c2ctuA_
not modelled
11.9
50
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 483;PDBTitle: solution structure of zinc finger domain from human zn2 finger protein 483
36 c2byvE_
not modelled
11.3
13
PDB header: regulationChain: E: PDB Molecule: rap guanine nucleotide exchange factor 4;PDBTitle: structure of the camp responsive exchange factor epac2 in2 its auto-inhibited state
37 d2cqna1
not modelled
11.3
27
Fold: Another 3-helical bundleSuperfamily: FF domainFamily: FF domain38 c1t39A_
not modelled
11.0
57
PDB header: transferase/dnaChain: A: PDB Molecule: methylated-dna--protein-cysteinePDBTitle: human o6-alkylguanine-dna alkyltransferase covalently2 crosslinked to dna
39 d1lj5a_
not modelled
10.8
9
Fold: SerpinsSuperfamily: SerpinsFamily: Serpins40 c3nwrA_
not modelled
10.7
16
PDB header: lyaseChain: A: PDB Molecule: a rubisco-like protein;PDBTitle: crystal structure of a rubisco-like protein from burkholderia fungorum
41 c3i71B_
not modelled
10.1
28
PDB header: unknown functionChain: B: PDB Molecule: ethanolamine utilization protein eutk;PDBTitle: ethanolamine utilization microcompartment shell subunit, eutk c-2 terminal domain
42 c3r4kD_
not modelled
10.1
16
PDB header: dna binding proteinChain: D: PDB Molecule: transcriptional regulator, iclr family;PDBTitle: crystal structure of a putative iclr transcriptional regulator2 (tm1040_3717) from silicibacter sp. tm1040 at 2.46 a resolution
43 c2ijeS_
not modelled
10.1
18
PDB header: signaling proteinChain: S: PDB Molecule: guanine nucleotide-releasing protein;PDBTitle: crystal structure of the cdc25 domain of rasgrf1
44 c2fyuE_
not modelled
10.1
64
PDB header: oxidoreductaseChain: E: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit,PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
45 c3bmxB_
not modelled
9.9
25
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized lipoprotein ybbd;PDBTitle: beta-n-hexosaminidase (ybbd) from bacillus subtilis
46 c3ndaA_
not modelled
9.9
16
PDB header: hydrolase inhibitorChain: A: PDB Molecule: serpin-2;PDBTitle: crystal structure of serpin from tick ixodes ricinus
47 d1whna_
not modelled
9.8
18
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)48 c3co7C_
not modelled
9.6
16
PDB header: transcription/dnaChain: C: PDB Molecule: forkhead box protein o1;PDBTitle: crystal structure of foxo1 dbd bound to dbe2 dna
49 d1seka_
not modelled
9.5
14
Fold: SerpinsSuperfamily: SerpinsFamily: Serpins50 d1dqna_
not modelled
9.4
14
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)51 c3ozqA_
not modelled
9.2
16
PDB header: hydrolase inhibitorChain: A: PDB Molecule: serpin48;PDBTitle: crystal structure of serpin48, which is a highly specific serpin in2 the insect tenebrio molitor
52 c1p84E_
not modelled
8.9
50
PDB header: oxidoreductaseChain: E: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfurPDBTitle: hdbt inhibited yeast cytochrome bc1 complex
53 d2qsba1
not modelled
8.8
17
Fold: Bromodomain-likeSuperfamily: Ta0600-likeFamily: Ta0600-like54 d2hfha_
not modelled
8.7
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain55 d1ykwa1
not modelled
8.7
14
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain56 c3c01H_
not modelled
8.5
20
PDB header: membrane protein, protein transportChain: H: PDB Molecule: surface presentation of antigens protein spas;PDBTitle: crystal structural of native spas c-terminal domain
57 c3psfA_
not modelled
8.0
19
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt6;PDBTitle: crystal structure of the spt6 core domain from saccharomyces2 cerevisiae, form spt6(236-1259)
58 c2l25A_
not modelled
7.9
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: np_888769.1
59 d8ruca1
not modelled
7.8
12
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain60 d1kcfa2
not modelled
7.7
15
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Mitochondrial resolvase ydc2 catalytic domain61 d1jm6a1
not modelled
7.7
19
Fold: Bromodomain-likeSuperfamily: alpha-ketoacid dehydrogenase kinase, N-terminal domainFamily: alpha-ketoacid dehydrogenase kinase, N-terminal domain62 c2h4qA_
not modelled
7.5
15
PDB header: hydrolase inhibitorChain: A: PDB Molecule: heterochromatin-associated protein ment;PDBTitle: crystal structure of a m-loop deletion variant of ment in2 the cleaved conformation
63 c3kcnA_
not modelled
7.4
5
PDB header: lyaseChain: A: PDB Molecule: adenylate cyclase homolog;PDBTitle: the crystal structure of adenylate cyclase from2 rhodopirellula baltica
64 c1mgtA_
not modelled
7.4
71
PDB header: transferaseChain: A: PDB Molecule: protein (o6-methylguanine-dna methyltransferase);PDBTitle: crystal structure of o6-methylguanine-dna methyltransferase from2 hyperthermophilic archaeon pyrococcus kodakaraensis strain kod1
65 c3t7yB_
not modelled
7.3
17
PDB header: protein transportChain: B: PDB Molecule: yop proteins translocation protein u;PDBTitle: structure of an autocleavage-inactive mutant of the cytoplasmic domain2 of ct091, the yscu homologue of chlamydia trachomatis
66 c2kk6A_
not modelled
7.3
22
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase fer;PDBTitle: solution structure of sh2 domain of proto-oncogene tyrosine-2 protein kinase fer from homo sapiens, northeast structural3 genomics consortium (nesg) target hr3461d
67 c3igzB_
not modelled
7.3
22
PDB header: isomeraseChain: B: PDB Molecule: cofactor-independent phosphoglycerate mutase;PDBTitle: crystal structures of leishmania mexicana phosphoglycerate2 mutase at low cobalt concentration
68 d1ej7l1
not modelled
7.3
13
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain69 d1svda1
not modelled
7.2
18
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain70 c2vh4B_
not modelled
7.2
13
PDB header: hydrolase inhibitorChain: B: PDB Molecule: tengpin;PDBTitle: structure of a loop c-sheet serpin polymer
71 d1d7ka2
not modelled
7.2
24
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain72 d3bzra1
not modelled
7.1
12
Fold: EscU C-terminal domain-likeSuperfamily: EscU C-terminal domain-likeFamily: EscU C-terminal domain-like73 c3bzrA_
not modelled
7.1
12
PDB header: membrane protein, protein transportChain: A: PDB Molecule: escu;PDBTitle: crystal structure of escu c-terminal domain with n262d mutation, space2 group p 41 21 2
74 c2fynO_
not modelled
7.1
50
PDB header: oxidoreductaseChain: O: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfurPDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
75 d3bpya1
not modelled
7.1
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain76 d1k9oi_
not modelled
7.0
14
Fold: SerpinsSuperfamily: SerpinsFamily: Serpins77 c1hleA_
not modelled
6.9
17
PDB header: hydrolase inhibitor(serine proteinase)Chain: A: PDB Molecule: horse leukocyte elastase inhibitor;PDBTitle: crystal structure of cleaved equine leucocyte elastase2 inhibitor determined at 1.95 angstroms resolution
78 c2imgA_
not modelled
6.9
33
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 23;PDBTitle: crystal structure of dual specificity protein phosphatase2 23 from homo sapiens in complex with ligand malate ion
79 d1kq8a_
not modelled
6.6
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain80 c1kq8A_
not modelled
6.6
27
PDB header: transcriptionChain: A: PDB Molecule: hepatocyte nuclear factor 3 forkhead homolog 1;PDBTitle: solution structure of winged helix protein hfh-1
81 c2hxhC_
not modelled
6.5
19
PDB header: transport proteinChain: C: PDB Molecule: kinesin-like protein kif1a;PDBTitle: kif1a head-microtubule complex structure in adp-form
82 c3euhB_
not modelled
6.5
18
PDB header: cell cycleChain: B: PDB Molecule: chromosome partition protein mukf;PDBTitle: crystal structure of the muke-mukf complex
83 c1sngA_
not modelled
6.4
22
PDB header: hydrolase inhibitorChain: A: PDB Molecule: cog4826: serine protease inhibitor;PDBTitle: structure of a thermophilic serpin in the native state
84 d1imva_
not modelled
6.3
19
Fold: SerpinsSuperfamily: SerpinsFamily: Serpins85 d2doea1
not modelled
6.2
14
Fold: Another 3-helical bundleSuperfamily: FF domainFamily: FF domain86 d2qlvb1
not modelled
6.0
36
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: AMPK-beta glycogen binding domain-like87 c3rgqA_
not modelled
6.0
22
PDB header: hydrolaseChain: A: PDB Molecule: protein-tyrosine phosphatase mitochondrial 1;PDBTitle: crystal structure of ptpmt1 in complex with pi(5)p
88 d1x38a1
not modelled
5.9
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: NagZ-like89 d1riea_
not modelled
5.9
64
Fold: ISP domainSuperfamily: ISP domainFamily: Rieske iron-sulfur protein (ISP)90 c3qfwB_
not modelled
5.8
20
PDB header: lyaseChain: B: PDB Molecule: ribulose-1,5-bisphosphate carboxylase/oxygenase largePDBTitle: crystal structure of rubisco-like protein from rhodopseudomonas2 palustris
91 c1t5cA_
not modelled
5.7
12
PDB header: contractile proteinChain: A: PDB Molecule: centromeric protein e;PDBTitle: crystal structure of the motor domain of human kinetochore2 protein cenp-e
92 d2z15a1
not modelled
5.7
10
Fold: BTG domain-likeSuperfamily: BTG domain-likeFamily: BTG domain-like93 c3fcnA_
not modelled
5.7
18
PDB header: unknown functionChain: A: PDB Molecule: an alpha-helical protein of unknown function (pfam01724);PDBTitle: crystal structure of an alpha-helical protein of unknown function2 (rru_a3208) from rhodospirillum rubrum atcc 11170 at 1.45 a3 resolution
94 d3e9va1
not modelled
5.6
14
Fold: BTG domain-likeSuperfamily: BTG domain-likeFamily: BTG domain-like95 d1sfea1
not modelled
5.6
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domainFamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domain96 c2k7rA_
not modelled
5.5
11
PDB header: replicationChain: A: PDB Molecule: primosomal protein dnai;PDBTitle: n-terminal domain of the bacillus subtilis helicase-loading2 protein dnai
97 c2ia2D_
not modelled
5.5
12
PDB header: transcriptionChain: D: PDB Molecule: putative transcriptional regulator;PDBTitle: the crystal structure of a putative transcriptional regulator rha061952 from rhodococcus sp. rha1
98 c2nvgA_
not modelled
5.4
45
PDB header: oxidoreductaseChain: A: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit;PDBTitle: soluble domain of rieske iron sulfur protein.
99 d2bsqe1
not modelled
5.4
29
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Trafficking protein A-like