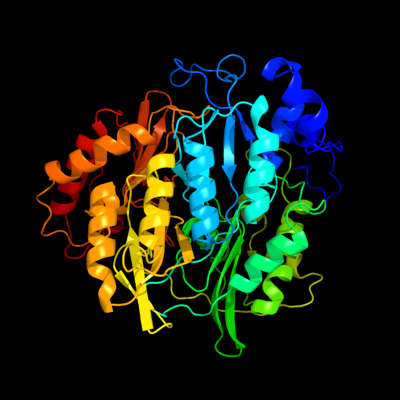
| 1 | c1zj8B_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable ferredoxin-dependent nitrite reductase nira;
PDBTitle: structure of mycobacterium tuberculosis nira protein
|
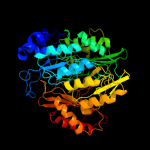
| 2 | c2akjA_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin--nitrite reductase, chloroplast;
PDBTitle: structure of spinach nitrite reductase
|
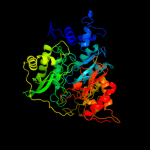
| 3 | c5aopA_
|
|
 |
100.0 |
100 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase hemoprotein;
PDBTitle: sulfite reductase structure reduced with crii edta, 5-coordinate2 siroheme, siroheme feii, [4fe-4s] +1
|
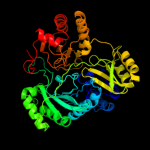
| 4 | c2v4jA_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase, dissimilatory-type subunit
PDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
|
| 5 | c2v4jE_
|
|
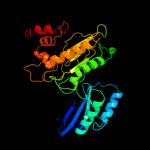 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:sulfite reductase, dissimilatory-type subunit
PDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
|
| 6 | c3c7bE_
|
|
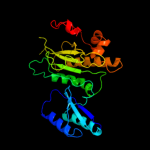 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:sulfite reductase, dissimilatory-type subunit beta;
PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
|
| 7 | c3c7bA_
|
|
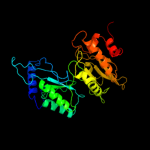 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase, dissimilatory-type subunit alpha;
PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
|
| 8 | d1aopa3
|
|
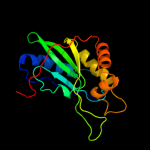 |
100.0 |
100 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 9 | d1aopa4
|
|
 |
100.0 |
100 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 10 | d1zj8a2
|
|
 |
100.0 |
19 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 11 | d2akja4
|
|
 |
100.0 |
27 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 12 | d1zj8a4
|
|
 |
100.0 |
25 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 13 | d2akja2
|
|
 |
100.0 |
24 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 14 | d1zj8a3
|
|
 |
100.0 |
25 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 15 | d3c7bb3
|
|
 |
100.0 |
16 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 16 | d2v4jb3
|
|
 |
100.0 |
16 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 17 | d3c7ba3
|
|
 |
100.0 |
18 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 18 | d2v4ja3
|
|
 |
100.0 |
14 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 19 | d2akja3
|
|
 |
100.0 |
22 |
Fold:Nitrite and sulphite reductase 4Fe-4S domain-like
Superfamily:Nitrite and sulphite reductase 4Fe-4S domain-like
Family:Nitrite and sulphite reductase 4Fe-4S domain-like |
| 20 | d3c7bb2
|
|
 |
99.8 |
11 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:DsrA/DsrB N-terminal-domain-like |
| 21 | d1zj8a1 |
|
not modelled |
99.8 |
15 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 22 | d1aopa1 |
|
not modelled |
99.8 |
98 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 23 | d2v4jb2 |
|
not modelled |
99.8 |
11 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:DsrA/DsrB N-terminal-domain-like |
| 24 | d1aopa2 |
|
not modelled |
99.7 |
100 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 25 | d2akja1 |
|
not modelled |
99.7 |
14 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 26 | c3noyA_ |
|
not modelled |
99.7 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: crystal structure of ispg (gcpe)
|
| 27 | d3c7ba2 |
|
not modelled |
98.9 |
18 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:DsrA/DsrB N-terminal-domain-like |
| 28 | c2y0fD_ |
|
not modelled |
98.4 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
|
| 29 | d2v4ja2 |
|
not modelled |
97.9 |
14 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:DsrA/DsrB N-terminal-domain-like |
| 30 | d1f37b_ |
|
not modelled |
78.7 |
22 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioredoxin-like 2Fe-2S ferredoxin |
| 31 | d2fug21 |
|
not modelled |
78.1 |
21 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:NQO2-like |
| 32 | c3nqoB_ |
|
not modelled |
73.0 |
11 |
PDB header:transcription
Chain: B: PDB Molecule:marr-family transcriptional regulator;
PDBTitle: crystal structure of a marr family transcriptional regulator (cd1569)2 from clostridium difficile 630 at 2.20 a resolution
|
| 33 | c2nyxB_ |
|
not modelled |
67.9 |
10 |
PDB header:transcription
Chain: B: PDB Molecule:probable transcriptional regulatory protein, rv1404;
PDBTitle: crystal structure of rv1404 from mycobacterium tuberculosis
|
| 34 | d1m2da_ |
|
not modelled |
61.4 |
28 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioredoxin-like 2Fe-2S ferredoxin |
| 35 | d3broa1 |
|
not modelled |
58.7 |
11 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 36 | c3nrvC_ |
|
not modelled |
58.5 |
15 |
PDB header:transcription regulator
Chain: C: PDB Molecule:putative transcriptional regulator (marr/emrr family);
PDBTitle: crystal structure of marr/emrr family transcriptional regulator from2 acinetobacter sp. adp1
|
| 37 | d1lnwa_ |
|
not modelled |
56.2 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 38 | c2nnnB_ |
|
not modelled |
54.4 |
22 |
PDB header:transcription
Chain: B: PDB Molecule:probable transcriptional regulator;
PDBTitle: crystal structure of probable transcriptional regulator from2 pseudomonas aeruginosa
|
| 39 | d1s3ja_ |
|
not modelled |
54.3 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 40 | d2etha1 |
|
not modelled |
48.0 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 41 | c3k0lA_ |
|
not modelled |
47.4 |
19 |
PDB header:transcription regulator
Chain: A: PDB Molecule:repressor protein;
PDBTitle: crystal structure of putative marr family transcriptional2 regulator from acinetobacter sp. adp
|
| 42 | c3g3zA_ |
|
not modelled |
47.4 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: the structure of nmb1585, a marr family regulator from neisseria2 meningitidis
|
| 43 | c3bpxB_ |
|
not modelled |
45.7 |
10 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator;
PDBTitle: crystal structure of marr
|
| 44 | d1ve4a1 |
|
not modelled |
44.8 |
13 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 45 | d2bv6a1 |
|
not modelled |
43.4 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 46 | d1p4xa2 |
|
not modelled |
41.8 |
7 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 47 | d1z7me1 |
|
not modelled |
40.8 |
7 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 48 | c3e6mD_ |
|
not modelled |
39.5 |
14 |
PDB header:transcription regulator
Chain: D: PDB Molecule:marr family transcriptional regulator;
PDBTitle: the crystal structure of a marr family transcriptional2 regulator from silicibacter pomeroyi dss.
|
| 49 | c3bjaA_ |
|
not modelled |
39.3 |
11 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, marr family, putative;
PDBTitle: crystal structure of putative marr-like transcription regulator2 (np_978771.1) from bacillus cereus atcc 10987 at 2.38 a resolution
|
| 50 | d1xdpa1 |
|
not modelled |
38.6 |
15 |
Fold:Spectrin repeat-like
Superfamily:PPK N-terminal domain-like
Family:PPK N-terminal domain-like |
| 51 | c3bj6B_ |
|
not modelled |
37.6 |
13 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of marr family transcription regulator sp03579
|
| 52 | c3ereD_ |
|
not modelled |
36.8 |
15 |
PDB header:dna binding protein/dna
Chain: D: PDB Molecule:arginine repressor;
PDBTitle: crystal structure of the arginine repressor protein from mycobacterium2 tuberculosis in complex with the dna operator
|
| 53 | c1b4aA_ |
|
not modelled |
36.4 |
7 |
PDB header:repressor
Chain: A: PDB Molecule:arginine repressor;
PDBTitle: structure of the arginine repressor from bacillus stearothermophilus
|
| 54 | d2a61a1 |
|
not modelled |
35.0 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 55 | d2fbha1 |
|
not modelled |
34.8 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 56 | d2fbia1 |
|
not modelled |
32.6 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 57 | d1nh8a1 |
|
not modelled |
31.4 |
11 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 58 | c1nh7A_ |
|
not modelled |
31.4 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:atp phosphoribosyltransferase;
PDBTitle: atp phosphoribosyltransferase (atp-prtase) from mycobacterium2 tuberculosis
|
| 59 | d1c0aa2 |
|
not modelled |
31.3 |
15 |
Fold:DCoH-like
Superfamily:GAD domain-like
Family:GAD domain |
| 60 | d1xxaa_ |
|
not modelled |
30.9 |
9 |
Fold:DCoH-like
Superfamily:C-terminal domain of arginine repressor
Family:C-terminal domain of arginine repressor |
| 61 | c3cagF_ |
|
not modelled |
29.8 |
15 |
PDB header:dna binding protein
Chain: F: PDB Molecule:arginine repressor;
PDBTitle: crystal structure of the oligomerization domain hexamer of the2 arginine repressor protein from mycobacterium tuberculosis in complex3 with 9 arginines.
|
| 62 | c2auvA_ |
|
not modelled |
28.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:potential nad-reducing hydrogenase subunit;
PDBTitle: solution structure of hndac : a thioredoxin-like [2fe-2s]2 ferredoxin involved in the nadp-reducing hydrogenase3 complex
|
| 63 | c2fxaB_ |
|
not modelled |
28.3 |
10 |
PDB header:transcription
Chain: B: PDB Molecule:protease production regulatory protein hpr;
PDBTitle: structure of the protease production regulatory protein hpr from2 bacillus subtilis.
|
| 64 | d1lj9a_ |
|
not modelled |
26.6 |
9 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 65 | d2o8ra1 |
|
not modelled |
26.6 |
17 |
Fold:Spectrin repeat-like
Superfamily:PPK N-terminal domain-like
Family:PPK N-terminal domain-like |
| 66 | c1r1gB_ |
|
not modelled |
26.3 |
57 |
PDB header:toxin
Chain: B: PDB Molecule:neurotoxin bmk37;
PDBTitle: crystal structure of the scorpion toxin bmbkttx1
|
| 67 | d1r1ga_ |
|
not modelled |
26.3 |
57 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Scorpion toxin-like
Family:Short-chain scorpion toxins |
| 68 | c1r1gA_ |
|
not modelled |
26.3 |
57 |
PDB header:toxin
Chain: A: PDB Molecule:neurotoxin bmk37;
PDBTitle: crystal structure of the scorpion toxin bmbkttx1
|
| 69 | d1b5ta_ |
|
not modelled |
26.1 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:FAD-linked oxidoreductase
Family:Methylenetetrahydrofolate reductase |
| 70 | d1olta_ |
|
not modelled |
25.9 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Oxygen-independent coproporphyrinogen III oxidase HemN |
| 71 | d2r48a1 |
|
not modelled |
24.3 |
38 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Fructose specific IIB subunit-like |
| 72 | c2qwwB_ |
|
not modelled |
24.0 |
16 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of multiple antibiotic-resistance repressor (marr)2 (yp_013417.1) from listeria monocytogenes 4b f2365 at 2.07 a3 resolution
|
| 73 | d1zhva2 |
|
not modelled |
23.9 |
9 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Atu0741-like |
| 74 | d1qrjb1 |
|
not modelled |
23.8 |
25 |
Fold:Acyl carrier protein-like
Superfamily:Retrovirus capsid dimerization domain-like
Family:Retrovirus capsid protein C-terminal domain |
| 75 | c3cdhB_ |
|
not modelled |
22.5 |
7 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of the marr family transcriptional regulator spo14532 from silicibacter pomeroyi dss-3
|
| 76 | c1p4xA_ |
|
not modelled |
21.9 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:staphylococcal accessory regulator a homologue;
PDBTitle: crystal structure of sars protein from staphylococcus aureus
|
| 77 | d1z91a1 |
|
not modelled |
21.8 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 78 | d2r4qa1 |
|
not modelled |
21.5 |
50 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Fructose specific IIB subunit-like |
| 79 | d1hsja1 |
|
not modelled |
20.6 |
7 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 80 | d1s4ea2 |
|
not modelled |
20.2 |
16 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Galactokinase |
| 81 | c2kyrA_ |
|
not modelled |
19.9 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:fructose-like phosphotransferase enzyme iib component 1;
PDBTitle: solution structure of enzyme iib subunit of pts system from2 escherichia coli k12. northeast structural genomics consortium target3 er315/ontario center for structural proteomics target ec0544
|
| 82 | c3lx4B_ |
|
not modelled |
19.6 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:fe-hydrogenase;
PDBTitle: stepwise [fefe]-hydrogenase h-cluster assembly revealed in the2 structure of hyda(deltaefg)
|
| 83 | d1t6sa1 |
|
not modelled |
19.3 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:ScpB/YpuH-like |
| 84 | d1jgsa_ |
|
not modelled |
19.1 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 85 | c2vd2A_ |
|
not modelled |
19.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:atp phosphoribosyltransferase;
PDBTitle: the crystal structure of hisg from b. subtilis
|
| 86 | d1piea2 |
|
not modelled |
19.0 |
16 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Galactokinase |
| 87 | c1xdoB_ |
|
not modelled |
19.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of escherichia coli polyphosphate kinase
|
| 88 | d2p5ma1 |
|
not modelled |
18.7 |
11 |
Fold:DCoH-like
Superfamily:C-terminal domain of arginine repressor
Family:C-terminal domain of arginine repressor |
| 89 | d1ffgb_ |
|
not modelled |
18.4 |
13 |
Fold:Ferredoxin-like
Superfamily:CheY-binding domain of CheA
Family:CheY-binding domain of CheA |
| 90 | c3cjnA_ |
|
not modelled |
18.4 |
16 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of transcriptional regulator, marr family, from2 silicibacter pomeroyi
|
| 91 | c3oopA_ |
|
not modelled |
18.2 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin2960 protein;
PDBTitle: the structure of a protein with unknown function from listeria innocua2 clip11262
|
| 92 | c1zhvA_ |
|
not modelled |
17.8 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein atu0741;
PDBTitle: x-ray crystal structure protein atu0741 from agobacterium tumefaciens.2 northeast structural genomics consortium target atr8.
|
| 93 | d1jvaa3 |
|
not modelled |
17.7 |
15 |
Fold:Homing endonuclease-like
Superfamily:Homing endonucleases
Family:Intein endonuclease |
| 94 | c3hrmA_ |
|
not modelled |
17.6 |
8 |
PDB header:transcription regulator
Chain: A: PDB Molecule:hth-type transcriptional regulator sarz;
PDBTitle: crystal structure of staphylococcus aureus protein sarz in sulfenic2 acid form
|
| 95 | d1l0wa2 |
|
not modelled |
17.2 |
16 |
Fold:DCoH-like
Superfamily:GAD domain-like
Family:GAD domain |
| 96 | d1we6a_ |
|
not modelled |
16.7 |
16 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 97 | d1a8oa_ |
|
not modelled |
15.6 |
33 |
Fold:Acyl carrier protein-like
Superfamily:Retrovirus capsid dimerization domain-like
Family:Retrovirus capsid protein C-terminal domain |
| 98 | c3v4gA_ |
|
not modelled |
15.4 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:arginine repressor;
PDBTitle: 1.60 angstrom resolution crystal structure of an arginine repressor2 from vibrio vulnificus cmcp6
|
| 99 | d2eiaa1 |
|
not modelled |
15.0 |
33 |
Fold:Acyl carrier protein-like
Superfamily:Retrovirus capsid dimerization domain-like
Family:Retrovirus capsid protein C-terminal domain |