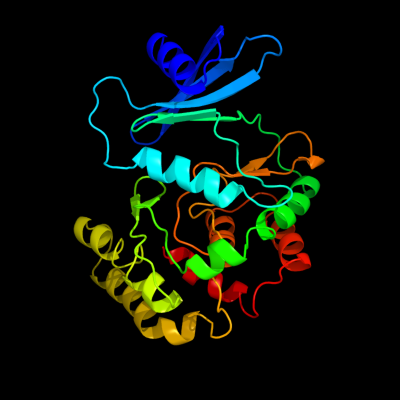
1 c3f7wA_
98.6
18
PDB header: transferaseChain: A: PDB Molecule: putative fructosamine-3-kinase;PDBTitle: crystal structure of putative fructosamine-3-kinase (yp_290396.1) from2 thermobifida fusca yx-er1 at 1.85 a resolution
2 c3csvA_
97.6
17
PDB header: transferaseChain: A: PDB Molecule: aminoglycoside phosphotransferase;PDBTitle: crystal structure of a putative aminoglycoside phosphotransferase2 (yp_614837.1) from silicibacter sp. tm1040 at 2.15 a resolution
3 d1j7la_
97.4
18
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: APH phosphotransferases4 c3lzhA_
97.0
12
PDB header: transferaseChain: A: PDB Molecule: aph(2")-id/aph(2")-iva;PDBTitle: crystal structure of aminoglycoside phosphotransferase aph(2")-2 id/aph(2")-iva
5 c3ovcA_
97.0
14
PDB header: transferase/antibioticChain: A: PDB Molecule: hygromycin-b 4-o-kinase;PDBTitle: crystal structure of aminoglycoside phosphotransferase aph(4)-ia
6 c3jr1A_
97.0
16
PDB header: transferaseChain: A: PDB Molecule: putative fructosamine-3-kinase;PDBTitle: crystal structure of putative fructosamine-3-kinase2 (yp_719053.1) from haemophilus somnus 129pt at 2.32 a3 resolution
7 d1nd4a_
96.9
18
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: APH phosphotransferases8 c3hamA_
96.2
14
PDB header: transferaseChain: A: PDB Molecule: aminoglycoside phosphotransferase;PDBTitle: structure of the gentamicin-aph(2")-iia complex
9 c3i0oA_
95.6
15
PDB header: transferaseChain: A: PDB Molecule: spectinomycin phosphotransferase;PDBTitle: crystal structure of spectinomycin phosphotransferase,2 aph(9)-ia, in complex with adp and spectinomcyin
10 c3attA_
94.7
17
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of rv3168 with atp
11 c3dxpA_
94.3
18
PDB header: transferaseChain: A: PDB Molecule: putative acyl-coa dehydrogenase;PDBTitle: crystal structure of a putative aminoglycoside phosphotransferase2 (reut_a1007) from ralstonia eutropha jmp134 at 2.32 a resolution
12 d2pula1
85.2
12
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: APH phosphotransferases13 c3r78B_
65.0
18
PDB header: transferaseChain: B: PDB Molecule: aminoglycoside 3'-phosphotransferase apha1-iab;PDBTitle: crystal structure of the aminoglycoside phosphotransferase aph(3')-ia,2 atp-bound
14 c2plyB_
57.1
23
PDB header: translation/rnaChain: B: PDB Molecule: selenocysteine-specific elongation factor;PDBTitle: structure of the mrna binding fragment of elongation factor2 selb in complex with secis rna.
15 d2v9va2
49.9
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: C-terminal fragment of elongation factor SelB16 c2xznH_
36.3
19
PDB header: ribosomeChain: H: PDB Molecule: ribosomal protein s8 containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
17 d1oy0a_
27.9
20
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Ketopantoate hydroxymethyltransferase PanB18 c2fs1A_
22.6
50
PDB header: protein bindingChain: A: PDB Molecule: psd-1;PDBTitle: solution structure of psd-1
19 c1s1hH_
22.1
18
PDB header: ribosomeChain: H: PDB Molecule: 40s ribosomal protein s22;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
20 d2gy9h1
19.9
26
Fold: Ribosomal protein S8Superfamily: Ribosomal protein S8Family: Ribosomal protein S821 d1gjsa_
not modelled
19.5
44
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Bacterial immunoglobulin/albumin-binding domainsFamily: GA module, an albumin-binding domain22 c1nyqA_
not modelled
18.2
32
PDB header: ligaseChain: A: PDB Molecule: threonyl-trna synthetase 1;PDBTitle: structure of staphylococcus aureus threonyl-trna synthetase2 complexed with an analogue of threonyl adenylate
23 d1i94h_
not modelled
17.7
24
Fold: Ribosomal protein S8Superfamily: Ribosomal protein S8Family: Ribosomal protein S824 d1an7a_
not modelled
16.7
24
Fold: Ribosomal protein S8Superfamily: Ribosomal protein S8Family: Ribosomal protein S825 d1seia_
not modelled
16.1
20
Fold: Ribosomal protein S8Superfamily: Ribosomal protein S8Family: Ribosomal protein S826 d1na6a2
not modelled
14.7
25
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Type II restriction endonuclease catalytic domain27 c2v9vA_
not modelled
14.4
24
PDB header: transcriptionChain: A: PDB Molecule: selenocysteine-specific elongation factor;PDBTitle: crystal structure of moorella thermoacetica selb(377-511)
28 c2pywA_
not modelled
13.7
15
PDB header: transferaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: structure of a. thaliana 5-methylthioribose kinase in complex with adp2 and mtr
29 c2q83A_
not modelled
13.0
13
PDB header: transferaseChain: A: PDB Molecule: ytaa protein;PDBTitle: crystal structure of ytaa (2635576) from bacillus subtilis at 2.50 a2 resolution
30 c1qf6A_
not modelled
12.7
20
PDB header: ligase/rnaChain: A: PDB Molecule: threonyl-trna synthetase;PDBTitle: structure of e. coli threonyl-trna synthetase complexed with its2 cognate trna
31 c3bbnH_
not modelled
12.2
22
PDB header: ribosomeChain: H: PDB Molecule: ribosomal protein s8;PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
32 c3ez4B_
not modelled
12.1
27
PDB header: transferaseChain: B: PDB Molecule: 3-methyl-2-oxobutanoate hydroxymethyltransferase;PDBTitle: crystal structure of 3-methyl-2-oxobutanoate2 hydroxymethyltransferase from burkholderia pseudomallei
33 d1m9fd_
not modelled
11.8
19
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain34 d1zyla1
not modelled
11.0
13
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: APH phosphotransferases35 d1ve2a1
not modelled
10.9
23
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase36 c2pqrD_
not modelled
10.8
88
PDB header: apoptosisChain: D: PDB Molecule: wd repeat protein ykr036c;PDBTitle: crystal structure of yeast fis1 complexed with a fragment of yeast2 caf4
37 d2ppqa1
not modelled
10.8
14
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: APH phosphotransferases38 c3dxqB_
not modelled
10.7
13
PDB header: transferaseChain: B: PDB Molecule: choline/ethanolamine kinase family protein;PDBTitle: crystal structure of choline/ethanolamine kinase family protein2 (np_106042.1) from mesorhizobium loti at 2.55 a resolution
39 c2f3iA_
not modelled
10.4
25
PDB header: transferaseChain: A: PDB Molecule: dna-directed rna polymerases i, ii, and iii 17.1PDBTitle: solution structure of a subunit of rna polymerase ii
40 d1m9dc_
not modelled
10.2
19
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain41 c2kdnA_
not modelled
9.6
36
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein pfe0790c;PDBTitle: solution structure of pfe0790c, a putative bola-like2 protein from the protozoan parasite plasmodium falciparum.
42 c2wlvA_
not modelled
9.6
22
PDB header: virus proteinChain: A: PDB Molecule: gag polyprotein;PDBTitle: structure of the n-terminal capsid domain of hiv-2
43 d1w0ba_
not modelled
9.0
43
Fold: Spectrin repeat-likeSuperfamily: Alpha-hemoglobin stabilizing protein AHSPFamily: Alpha-hemoglobin stabilizing protein AHSP44 d1nyra4
not modelled
9.0
30
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain45 c3cf6E_
not modelled
8.9
39
PDB header: signaling protein/gtp-binding proteinChain: E: PDB Molecule: rap guanine nucleotide exchange factor (gef) 4;PDBTitle: structure of epac2 in complex with cyclic-amp and rap
46 c3tr7A_
not modelled
8.7
32
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: structure of a uracil-dna glycosylase (ung) from coxiella burnetii
47 d1upka_
not modelled
8.6
21
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: Mo25 protein48 c1na6B_
not modelled
8.5
25
PDB header: hydrolaseChain: B: PDB Molecule: restriction endonuclease ecorii;PDBTitle: crystal structure of restriction endonuclease ecorii mutant2 r88a
49 d1m3ua_
not modelled
7.9
11
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Ketopantoate hydroxymethyltransferase PanB50 d2cqqa1
not modelled
7.6
40
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain51 c2f5xC_
not modelled
7.4
21
PDB header: transport proteinChain: C: PDB Molecule: bugd;PDBTitle: structure of periplasmic binding protein bugd
52 d1z8ua1
not modelled
7.2
43
Fold: Spectrin repeat-likeSuperfamily: Alpha-hemoglobin stabilizing protein AHSPFamily: Alpha-hemoglobin stabilizing protein AHSP53 c3lyrA_
not modelled
6.9
47
PDB header: transcription activatorChain: A: PDB Molecule: transcription factor coe1;PDBTitle: human early b-cell factor 1 (ebf1) dna-binding domain
54 c3cdxB_
not modelled
6.4
28
PDB header: hydrolaseChain: B: PDB Molecule: succinylglutamatedesuccinylase/aspartoacylase;PDBTitle: crystal structure of2 succinylglutamatedesuccinylase/aspartoacylase from3 rhodobacter sphaeroides
55 d1ouoa_
not modelled
6.4
25
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: Endonuclease I56 d1l9bh2
not modelled
5.6
100
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region57 c2owrD_
not modelled
5.2
6
PDB header: hydrolaseChain: D: PDB Molecule: uracil-dna glycosylase;PDBTitle: crystal structure of vaccinia virus uracil-dna glycosylase
58 d2f1fa1
not modelled
5.1
33
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like59 d2fgca2
not modelled
5.1
28
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like