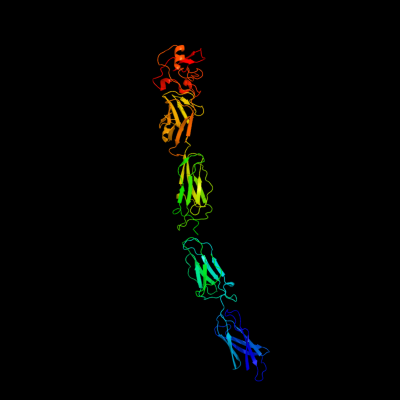1 c1cwvA_
100.0
23
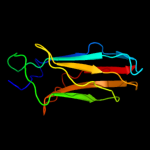
PDB header: structural proteinChain: A: PDB Molecule: invasin;PDBTitle: crystal structure of invasin: a bacterial integrin-binding protein
2 c1e5uI_
99.9
23
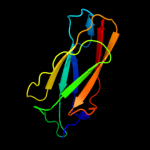
PDB header: intiminChain: I: PDB Molecule: intimin;PDBTitle: nmr representative structure of intimin-190 (int190) from2 enteropathogenic e. coli
3 c1f00I_
99.6
18
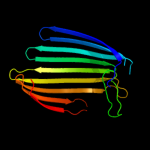
PDB header: cell adhesionChain: I: PDB Molecule: intimin;PDBTitle: crystal structure of c-terminal 282-residue fragment of2 enteropathogenic e. coli intimin
4 d1cwva4
98.8
34
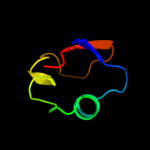
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments5 c2l9yA_
98.0
14
PDB header: sugar binding proteinChain: A: PDB Molecule: cvnh-lysm lectin;PDBTitle: solution structure of the mocvnh-lysm module from the rice blast2 fungus magnaporthe oryzae protein (mgg_03307)
6 c2djpA_
98.0
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein sb145;PDBTitle: the solution structure of the lysm domain of human2 hypothetical protein sb145
7 d1e0ga_
97.8
25
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain8 d1cwva3
97.7
22
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments9 d1cwva2
97.5
23
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments10 d2zfga1
97.5
12
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin11 d1y7ma2
97.5
24
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain12 d1phoa_
97.3
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin13 d1cwva5
97.2
23
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Invasin/intimin cell-adhesion fragment, C-terminal domain14 c2gu1A_
97.0
14
PDB header: hydrolaseChain: A: PDB Molecule: zinc peptidase;PDBTitle: crystal structure of a zinc containing peptidase from2 vibrio cholerae
15 c3nb3C_
96.8
19
PDB header: virusChain: C: PDB Molecule: outer membrane protein a;PDBTitle: the host outer membrane proteins ompa and ompc are packed at specific2 sites in the shigella phage sf6 virion as structural components
16 c2qomB_
96.8
11
PDB header: hydrolaseChain: B: PDB Molecule: serine protease espp;PDBTitle: the crystal structure of the e.coli espp autotransporter beta-domain.
17 c2k0lA_
96.6
19
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr structure of the transmembrane domain of the outer2 membrane protein a from klebsiella pneumoniae in dhpc3 micelles.
18 d1cwva1
96.5
25
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments19 c3aehB_
96.5
9
PDB header: hydrolaseChain: B: PDB Molecule: hemoglobin-binding protease hbp autotransporter;PDBTitle: integral membrane domain of autotransporter hbp
20 d1osma_
96.4
18
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin21 d1t16a_
not modelled
96.4
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Outer membrane protein transport protein22 c1y7mB_
not modelled
96.4
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein bsu14040;PDBTitle: crystal structure of the b. subtilis ykud protein at 2 a2 resolution
23 d1qj8a_
not modelled
96.4
15
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein24 c2x4mD_
not modelled
96.2
12
PDB header: hydrolaseChain: D: PDB Molecule: coagulase/fibrinolysin;PDBTitle: yersinia pestis plasminogen activator pla
25 d1f00i1
96.1
27
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments26 c3qq2C_
not modelled
96.1
12
PDB header: membrane protein/protein transportChain: C: PDB Molecule: brka autotransporter;PDBTitle: crystal structure of the beta domain of the bordetella autotransporter2 brka
27 d1qjpa_
not modelled
96.0
19
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein28 c3bryB_
not modelled
96.0
13
PDB header: transport proteinChain: B: PDB Molecule: tbux;PDBTitle: crystal structure of the ralstonia pickettii toluene2 transporter tbux
29 c3pdgA_
95.9
13
PDB header: unknown functionChain: A: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
30 c3qraA_
not modelled
95.8
14
PDB header: cell invasionChain: A: PDB Molecule: attachment invasion locus protein;PDBTitle: the crystal structure of ail, the attachment invasion locus protein of2 yersinia pestis
31 c2f1tB_
not modelled
95.8
13
PDB header: membrane proteinChain: B: PDB Molecule: outer membrane protein w;PDBTitle: outer membrane protein ompw
32 c3kvnA_
not modelled
95.6
10
PDB header: hydrolaseChain: A: PDB Molecule: esterase esta;PDBTitle: crystal structure of the full-length autotransporter esta from2 pseudomonas aeruginosa
33 c2jmmA_
not modelled
95.4
13
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr solution structure of a minimal transmembrane beta-2 barrel platform protein
34 c3pe9C_
95.0
15
PDB header: unknown functionChain: C: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
35 c3pe9A_
95.0
15
PDB header: unknown functionChain: A: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
36 d1g90a_
not modelled
94.8
16
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein37 c3pddA_
not modelled
94.3
12
PDB header: unknown functionChain: A: PDB Molecule: glycoside hydrolase, family 9;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
38 c3pe9D_
not modelled
94.0
13
PDB header: unknown functionChain: D: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
39 c2x27X_
not modelled
93.4
13
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein oprg;PDBTitle: crystal structure of the outer membrane protein oprg from2 pseudomonas aeruginosa
40 c3nsgA_
not modelled
92.9
18
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein f;PDBTitle: crystal structure of ompf, an outer membrane protein from salmonella2 typhi
41 c3brzA_
not modelled
92.5
14
PDB header: transport proteinChain: A: PDB Molecule: todx;PDBTitle: crystal structure of the pseudomonas putida toluene2 transporter todx
42 d1uynx_
not modelled
91.8
10
Fold: Transmembrane beta-barrelsSuperfamily: AutotransporterFamily: Autotransporter43 d3prna_
not modelled
91.8
14
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin44 c3pe9B_
not modelled
91.8
17
PDB header: unknown functionChain: B: PDB Molecule: fibronectin(iii)-like module;PDBTitle: structures of clostridium thermocellum cbha fibronectin(iii)-like2 modules
45 d1i78a_
not modelled
91.3
10
Fold: Transmembrane beta-barrelsSuperfamily: OMPT-likeFamily: Outer membrane protease OMPT46 d2pora_
not modelled
91.3
9
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin47 c2vqiA_
not modelled
91.1
15
PDB header: transportChain: A: PDB Molecule: outer membrane usher protein papc;PDBTitle: structure of the p pilus usher (papc) translocation pore
48 c3sljA_
not modelled
90.7
13
PDB header: protein transportChain: A: PDB Molecule: serine protease espp;PDBTitle: pre-cleavage structure of the autotransporter espp - n1023a mutant
49 d1p4ta_
not modelled
90.3
12
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein50 d2fgqx1
not modelled
89.4
17
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin51 c2o4vA_
not modelled
87.3
12
PDB header: membrane proteinChain: A: PDB Molecule: porin p;PDBTitle: an arginine ladder in oprp mediates phosphate specific transfer across2 the outer membrane
52 c3a2rX_
not modelled
81.8
10
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein ii;PDBTitle: crystal structure of outer membrane protein porb from neisseria2 meningitidis
53 d1pama3
not modelled
80.4
30
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain54 d1ja3a_
not modelled
78.4
16
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain55 d1qhoa3
not modelled
77.8
19
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain56 d1cyga3
not modelled
76.3
33
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain57 d3bmva3
not modelled
75.9
33
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain58 d1rdl1_
not modelled
75.8
17
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain59 d1cxla3
not modelled
75.8
30
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain60 d1byfa_
not modelled
75.7
29
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain61 d1cgta3
not modelled
75.6
33
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain62 d2afpa_
not modelled
75.6
14
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain63 c3c8jB_
not modelled
75.5
16
PDB header: immune systemChain: B: PDB Molecule: natural killer cell receptor ly49c;PDBTitle: the crystal structure of natural killer cell receptor ly49c
64 c2wjqA_
not modelled
75.2
9
PDB header: transport proteinChain: A: PDB Molecule: probable n-acetylneuraminic acid outer membrane channelPDBTitle: nanc porin structure in hexagonal crystal form.
65 c2lhfA_
not modelled
74.9
14
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein h1;PDBTitle: solution structure of outer membrane protein h (oprh) from p.2 aeruginosa in dhpc micelles
66 c3k7bA_
not modelled
74.8
21
PDB header: viral proteinChain: A: PDB Molecule: protein a33;PDBTitle: the structure of the poxvirus a33 protein reveals a dimer of unique c-2 type lectin-like domains.
67 c2dcjA_
not modelled
74.2
25
PDB header: hydrolaseChain: A: PDB Molecule: xylanase j;PDBTitle: a two-domain structure of alkaliphilic xynj from bacillus sp. 41m-1
68 c3dwoX_
not modelled
73.5
11
PDB header: membrane proteinChain: X: PDB Molecule: probable outer membrane protein;PDBTitle: crystal structure of a pseudomonas aeruginosa fadl homologue
69 d1wk1a_
not modelled
72.6
24
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain70 c1eslA_
not modelled
72.1
28
PDB header: cell adhesion proteinChain: A: PDB Molecule: human e-selectin;PDBTitle: insight into e-selectin(slash)ligand interaction from the2 crystal structure and mutagenesis of the lec(slash)egf3 domains
71 d1l0qa1
not modelled
71.2
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PKD domainFamily: PKD domain72 d3c8ja1
not modelled
70.8
16
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain73 c2bpdB_
70.7
17
PDB header: receptorChain: B: PDB Molecule: dectin-1;PDBTitle: structure of murine dectin-1
74 d1tlya_
not modelled
69.5
23
Fold: Transmembrane beta-barrelsSuperfamily: Tsx-like channelFamily: Tsx-like channel75 d1yu3a2
not modelled
68.7
19
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Mtd variable domain76 c2lfcA_
not modelled
68.5
31
PDB header: oxidoreductaseChain: A: PDB Molecule: fumarate reductase, flavoprotein subunit;PDBTitle: solution nmr structure of fumarate reductase flavoprotein subunit from2 lactobacillus plantarum, northeast structural genomics consortium3 target lpr145j
77 c2iwvD_
not modelled
67.7
21
PDB header: ion channelChain: D: PDB Molecule: outer membrane protein g;PDBTitle: structure of the monomeric outer membrane porin ompg in the2 open and closed conformation
78 c2zwkA_
not modelled
66.5
30
PDB header: cell adhesionChain: A: PDB Molecule: intimin;PDBTitle: crystal structure of intimin-tir90 complex
79 d1y4ja1
not modelled
66.4
23
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Sulfatase-modifying factor-like80 c2q17C_
not modelled
66.3
17
PDB header: unknown functionChain: C: PDB Molecule: formylglycine generating enzyme;PDBTitle: formylglycine generating enzyme from streptomyces coelicolor
81 d1yu0a2
not modelled
65.7
19
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Mtd variable domain82 d1z70x1
not modelled
65.1
20
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Sulfatase-modifying factor-like83 d2msba_
not modelled
64.2
26
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain84 d1rjha_
not modelled
63.9
10
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain85 c1y1fX_
not modelled
61.5
23
PDB header: oxidoreductaseChain: X: PDB Molecule: c-alpha-formyglycine-generating enzyme;PDBTitle: human formylglycine generating enzyme with cysteine sulfenic acid
86 d2gufa1
not modelled
61.2
11
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Ligand-gated protein channel87 c2kzwA_
not modelled
60.6
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of q8psa4 from methanosarcina mazei, northeast2 structural genomics consortium target mar143a
88 c2zibA_
not modelled
59.7
15
PDB header: antifreeze proteinChain: A: PDB Molecule: type ii antifreeze protein;PDBTitle: crystal structure analysis of calcium-independent type ii2 antifreeze protein
89 d1g1ta1
not modelled
58.9
22
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain90 c1hupA_
not modelled
58.5
13
PDB header: c-type lectinChain: A: PDB Molecule: mannose-binding protein;PDBTitle: human mannose binding protein carbohydrate recognition domain2 trimerizes through a triple alpha-helical coiled-coil
91 c2k53A_
not modelled
58.2
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: a3dk08 protein;PDBTitle: nmr solution structure of a3dk08 protein from clostridium2 thermocellum: northeast structural genomics consortium3 target cmr9
92 c3dbzB_
not modelled
58.1
16
PDB header: sugar binding proteinChain: B: PDB Molecule: pulmonary surfactant-associated protein d;PDBTitle: human surfactant protein d
93 d1g1sa1
not modelled
57.7
9
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain94 d1r13a1
not modelled
57.6
9
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain95 d1xpha1
not modelled
57.5
6
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain96 c2iouC_
not modelled
57.1
17
PDB header: viral protein/membrane proteinChain: C: PDB Molecule: major tropism determinant p1;PDBTitle: major tropism determinant p1 (mtd-p1) variant complexed with2 bordetella brochiseptica virulence factor pertactin extracellular3 domain (prn-e).
97 d1rr7a_
not modelled
56.7
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Middle operon regulator, Mor98 c1rr7A_
not modelled
56.7
14
PDB header: transcriptionChain: A: PDB Molecule: middle operon regulator;PDBTitle: crystal structure of the middle operon regulator protein of2 bacteriophage mu
99 c1fm5A_
not modelled
56.4
23
PDB header: immune systemChain: A: PDB Molecule: early activation antigen cd69;PDBTitle: crystal structure of human cd69
100 c3cfwA_
not modelled
56.2
30
PDB header: cell adhesionChain: A: PDB Molecule: l-selectin;PDBTitle: l-selectin lectin and egf domains
101 c2yrlA_
not modelled
54.2
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: kiaa1837 protein;PDBTitle: solution structure of the pkd domain from kiaa 1837 protein
102 c1l0qC_
not modelled
53.8
17
PDB header: protein bindingChain: C: PDB Molecule: surface layer protein;PDBTitle: tandem yvtn beta-propeller and pkd domains from an archaeal surface2 layer protein
103 c2elhA_
not modelled
53.5
33
PDB header: dna binding proteinChain: A: PDB Molecule: cg11849-pa;PDBTitle: solution structure of the cenp-b n-terminal dna-binding2 domain of fruit fly distal antenna cg11849-pa
104 c3rfzB_
not modelled
52.9
12
PDB header: cell adhesion/transport/chaperoneChain: B: PDB Molecule: outer membrane usher protein, type 1 fimbrial synthesis;PDBTitle: crystal structure of the fimd usher bound to its cognate fimc:fimh2 substrate
105 d1egga_
not modelled
52.5
14
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain106 d1fepa_
not modelled
52.3
12
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Ligand-gated protein channel107 c3m9zA_
not modelled
52.1
7
PDB header: signaling proteinChain: A: PDB Molecule: killer cell lectin-like receptor subfamily b member 1a;PDBTitle: crystal structure of extracellular domain of mouse nkr-p1a
108 c2c5uA_
51.7
30
PDB header: ligaseChain: A: PDB Molecule: rna ligase;PDBTitle: t4 rna ligase (rnl1) crystal structure
109 d1by5a_
not modelled
51.7
11
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Ligand-gated protein channel110 c2py2E_
not modelled
51.3
13
PDB header: antifreeze proteinChain: E: PDB Molecule: antifreeze protein type ii;PDBTitle: structure of herring type ii antifreeze protein
111 d1f00i3
not modelled
51.2
30
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Invasin/intimin cell-adhesion fragment, C-terminal domain112 c1kwwC_
not modelled
50.3
16
PDB header: immune system, sugar binding proteinChain: C: PDB Molecule: mannose-binding protein a;PDBTitle: rat mannose protein a complexed with a-me-fuc.
113 d1o75a1
not modelled
49.7
35
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Tp47 lipoprotein, middle and C-terminal domainsFamily: Tp47 lipoprotein, middle and C-terminal domains114 d1y0pa3
not modelled
48.9
17
Fold: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainSuperfamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain115 c3orjA_
not modelled
48.8
17
PDB header: sugar binding proteinChain: A: PDB Molecule: sugar-binding protein;PDBTitle: crystal structure of a sugar-binding protein (bacova_04391) from2 bacteroides ovatus at 2.16 a resolution
116 d1fifa1
not modelled
48.7
27
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain117 c2ostC_
not modelled
47.7
40
PDB header: hydrolase/dnaChain: C: PDB Molecule: putative endonuclease;PDBTitle: the structure of a bacterial homing endonuclease : i-ssp6803i
118 d2jn6a1
not modelled
46.6
30
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Cgl2762-like119 c1g1qD_
not modelled
45.9
21
PDB header: immune system, membrane proteinChain: D: PDB Molecule: p-selectin;PDBTitle: crystal structure of p-selectin lectin/egf domains
120 c2pmzV_
not modelled
45.6
18
PDB header: translation, transferaseChain: V: PDB Molecule: dna-directed rna polymerase subunit h;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus