| 1 | d2f9ya1
|
|
 |
100.0 |
99 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 2 | c2f9iC_
|
|
 |
100.0 |
54 |
PDB header:transferase
Chain: C: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl
PDBTitle: crystal structure of the carboxyltransferase subunit of acc2 from staphylococcus aureus
|
| 3 | c3glmD_
|
|
 |
100.0 |
15 |
PDB header:lyase
Chain: D: PDB Molecule:glutaconyl-coa decarboxylase subunit a;
PDBTitle: glutaconyl-coa decarboxylase a subunit from clostridium2 symbiosum co-crystallized with crotonyl-coa
|
| 4 | c1on3E_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: E: PDB Molecule:methylmalonyl-coa carboxyltransferase 12s
PDBTitle: transcarboxylase 12s crystal structure: hexamer assembly2 and substrate binding to a multienzyme core (with3 methylmalonyl-coenzyme a and methylmalonic acid bound)
|
| 5 | c1x0uB_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:hypothetical methylmalonyl-coa decarboxylase alpha subunit;
PDBTitle: crystal structure of the carboxyl transferase subunit of putative pcc2 of sulfolobus tokodaii
|
| 6 | c2a7sD_
|
|
 |
100.0 |
21 |
PDB header:ligase
Chain: D: PDB Molecule:probable propionyl-coa carboxylase beta chain 5;
PDBTitle: crystal structure of the acyl-coa carboxylase, accd5, from2 mycobacterium tuberculosis
|
| 7 | c1vrgE_
|
|
 |
100.0 |
22 |
PDB header:ligase
Chain: E: PDB Molecule:propionyl-coa carboxylase, beta subunit;
PDBTitle: crystal structure of propionyl-coa carboxylase, beta subunit (tm0716)2 from thermotoga maritima at 2.30 a resolution
|
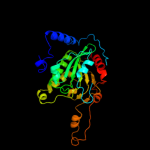
| 8 | c3n6rF_
|
|
 |
100.0 |
20 |
PDB header:ligase
Chain: F: PDB Molecule:propionyl-coa carboxylase, beta subunit;
PDBTitle: crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
|
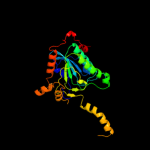
| 9 | d1pixa3
|
|
 |
100.0 |
16 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
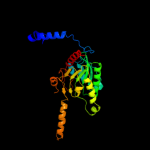
| 10 | c3u9rB_
|
|
 |
100.0 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:methylcrotonyl-coa carboxylase, beta-subunit;
PDBTitle: crystal structure of p. aeruginosa 3-methylcrotonyl-coa carboxylase2 (mcc), beta subunit
|
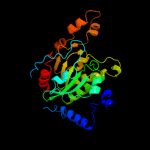
| 11 | c1xnwD_
|
|
 |
100.0 |
21 |
PDB header:ligase
Chain: D: PDB Molecule:propionyl-coa carboxylase complex b subunit;
PDBTitle: acyl-coa carboxylase beta subunit from s. coelicolor (pccb),2 apo form #2, mutant d422i
|
| 12 | c1pixB_
|
|
 |
100.0 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:glutaconyl-coa decarboxylase a subunit;
PDBTitle: crystal structure of the carboxyltransferase subunit of the2 bacterial ion pump glutaconyl-coenzyme a decarboxylase
|
| 13 | d2a7sa2
|
|
 |
100.0 |
25 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 14 | d1on3a2
|
|
 |
100.0 |
24 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 15 | d1xnya2
|
|
 |
100.0 |
22 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 16 | d1vrga2
|
|
 |
100.0 |
26 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 17 | d1pixa2
|
|
 |
100.0 |
15 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 18 | d1on3a1
|
|
 |
100.0 |
19 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 19 | d1vrga1
|
|
 |
100.0 |
14 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 20 | d1xnya1
|
|
 |
100.0 |
20 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 21 | d2a7sa1 |
|
not modelled |
100.0 |
19 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 22 | c2f9yB_ |
|
not modelled |
100.0 |
20 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl transferase subunit
PDBTitle: the crystal structure of the carboxyltransferase subunit of acc from2 escherichia coli
|
| 23 | d2f9yb1 |
|
not modelled |
100.0 |
20 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 24 | c2f9iD_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl
PDBTitle: crystal structure of the carboxyltransferase subunit of acc2 from staphylococcus aureus
|
| 25 | c2x24B_ |
|
not modelled |
100.0 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coa carboxylase;
PDBTitle: bovine acc2 ct domain in complex with inhibitor
|
| 26 | c3ff6D_ |
|
not modelled |
100.0 |
18 |
PDB header:ligase
Chain: D: PDB Molecule:acetyl-coa carboxylase 2;
PDBTitle: human acc2 ct domain with cp-640186
|
| 27 | c1od4C_ |
|
not modelled |
100.0 |
14 |
PDB header:ligase
Chain: C: PDB Molecule:acetyl-coenzyme a carboxylase;
PDBTitle: acetyl-coa carboxylase carboxyltransferase domain
|
| 28 | c1uytC_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: C: PDB Molecule:acetyl-coa carboxylase;
PDBTitle: acetyl-coa carboxylase carboxyltransferase domain
|
| 29 | c3h0jA_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:acetyl-coa carboxylase;
PDBTitle: crystal structure of the carboxyltransferase domain of2 acetyl-coenzyme a carboxylase in complex with compound 2
|
| 30 | d1uyra2 |
|
not modelled |
99.9 |
18 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 31 | d1uyra1 |
|
not modelled |
99.9 |
17 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 32 | c3r6hA_ |
|
not modelled |
99.2 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase, echa3;
PDBTitle: crystal structure of an enoyl-coa hydratase (echa3) from mycobacterium2 marinum
|
| 33 | c2fbmB_ |
|
not modelled |
99.2 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:y chromosome chromodomain protein 1, telomeric isoform b;
PDBTitle: acetyltransferase domain of cdy1
|
| 34 | d2a7ka1 |
|
not modelled |
99.2 |
11 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 35 | c3peaD_ |
|
not modelled |
99.1 |
10 |
PDB header:isomerase
Chain: D: PDB Molecule:enoyl-coa hydratase/isomerase family protein;
PDBTitle: crystal structure of enoyl-coa hydratase from bacillus anthracis str.2 'ames ancestor'
|
| 36 | c2q35A_ |
|
not modelled |
99.1 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:curf;
PDBTitle: crystal structure of the y82f variant of ech2 decarboxylase domain of2 curf from lyngbya majuscula
|
| 37 | d2fw2a1 |
|
not modelled |
99.1 |
16 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 38 | c2f6qA_ |
|
not modelled |
99.1 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:peroxisomal 3,2-trans-enoyl-coa isomerase;
PDBTitle: the crystal structure of human peroxisomal delta3, delta2 enoyl coa2 isomerase (peci)
|
| 39 | c3fduF_ |
|
not modelled |
99.1 |
18 |
PDB header:isomerase
Chain: F: PDB Molecule:putative enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of a putative enoyl-coa hydratase/isomerase from2 acinetobacter baumannii
|
| 40 | d1xx4a_ |
|
not modelled |
99.1 |
15 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 41 | c3i47A_ |
|
not modelled |
99.1 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl coa hydratase/isomerase (crotonase);
PDBTitle: crystal structure of putative enoyl coa hydratase/isomerase2 (crotonase) from legionella pneumophila subsp. pneumophila str.3 philadelphia 1
|
| 42 | c2ej5B_ |
|
not modelled |
99.1 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:enoyl-coa hydratase subunit ii;
PDBTitle: crystal structure of gk2038 protein (enoyl-coa hydratase subunit ii)2 from geobacillus kaustophilus
|
| 43 | d1nzya_ |
|
not modelled |
99.1 |
15 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 44 | c3rrvC_ |
|
not modelled |
99.1 |
17 |
PDB header:isomerase
Chain: C: PDB Molecule:enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of an enoyl-coa hydratase/isomerase from2 mycobacterium paratuberculosis
|
| 45 | c3lkeA_ |
|
not modelled |
99.1 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of enoyl-coa hydratase from bacillus2 halodurans
|
| 46 | c2vx2D_ |
|
not modelled |
99.1 |
15 |
PDB header:isomerase
Chain: D: PDB Molecule:enoyl-coa hydratase domain-containing protein 3;
PDBTitle: crystal structure of human enoyl coenzyme a hydratase2 domain-containing protein 3 (echdc3)
|
| 47 | d1wz8a1 |
|
not modelled |
99.0 |
16 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 48 | d2f6qa1 |
|
not modelled |
99.0 |
20 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 49 | c3hrxD_ |
|
not modelled |
99.0 |
20 |
PDB header:lyase
Chain: D: PDB Molecule:probable enoyl-coa hydratase;
PDBTitle: crystal structure of phenylacetic acid degradation protein paag
|
| 50 | c3mybA_ |
|
not modelled |
99.0 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of enoyl-coa hydratase mycobacterium smegmatis
|
| 51 | d1uiya_ |
|
not modelled |
99.0 |
20 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 52 | c3oc7A_ |
|
not modelled |
99.0 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of an enoyl-coa hydratase from mycobacterium avium
|
| 53 | c3kqfC_ |
|
not modelled |
99.0 |
14 |
PDB header:isomerase
Chain: C: PDB Molecule:enoyl-coa hydratase/isomerase family protein;
PDBTitle: 1.8 angstrom resolution crystal structure of enoyl-coa hydratase from2 bacillus anthracis.
|
| 54 | c3isaA_ |
|
not modelled |
99.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of putative enoyl-coa hydratase/isomerase from2 bordetella parapertussis
|
| 55 | d1dcia_ |
|
not modelled |
99.0 |
16 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 56 | c2qq3F_ |
|
not modelled |
99.0 |
12 |
PDB header:lyase
Chain: F: PDB Molecule:enoyl-coa hydratase subunit i;
PDBTitle: crystal structure of enoyl-coa hydrates subunit i (gk_2039)2 other form from geobacillus kaustophilus hta426
|
| 57 | c3q1tB_ |
|
not modelled |
99.0 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of enoyl-coa hydratase from mycobacterium avium
|
| 58 | c3l3sF_ |
|
not modelled |
99.0 |
14 |
PDB header:isomerase
Chain: F: PDB Molecule:enoyl-coa hydratase/isomerase family protein;
PDBTitle: crystal structure of an enoyl-coa hydrotase/isomerase family2 protein from silicibacter pomeroyi
|
| 59 | d1sg4a1 |
|
not modelled |
99.0 |
14 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 60 | c3omeE_ |
|
not modelled |
99.0 |
16 |
PDB header:lyase
Chain: E: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of a probable enoyl-coa hydratase from mycobacterium2 smegmatis
|
| 61 | c3g64A_ |
|
not modelled |
99.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:putative enoyl-coa hydratase;
PDBTitle: crystal structure of putative enoyl-coa hydratase from streptomyces2 coelicolor a3(2)
|
| 62 | d1hzda_ |
|
not modelled |
99.0 |
14 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 63 | c3njbA_ |
|
not modelled |
98.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of enoyl-coa hydratase from mycobacterium smegmatis,2 iodide soak
|
| 64 | c3moyA_ |
|
not modelled |
98.9 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:probable enoyl-coa hydratase;
PDBTitle: crystal structure of probable enoyl-coa hydratase from mycobacterium2 smegmatis
|
| 65 | c3p5mB_ |
|
not modelled |
98.9 |
21 |
PDB header:isomerase
Chain: B: PDB Molecule:enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of an enoyl-coa hydratase/isomerase from2 mycobacterium avium
|
| 66 | c3rsiA_ |
|
not modelled |
98.9 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:putative enoyl-coa hydratase/isomerase;
PDBTitle: the structure of a putative enoyl-coa hydratase/isomerase from2 mycobacterium abscessus atcc 19977 / dsm 44196
|
| 67 | c2ppyE_ |
|
not modelled |
98.9 |
14 |
PDB header:lyase
Chain: E: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of enoyl-coa hydrates (gk_1992) from geobacillus2 kaustophilus hta426
|
| 68 | d1pjha_ |
|
not modelled |
98.9 |
16 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 69 | c2iexA_ |
|
not modelled |
98.9 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxynapthoic acid synthetase;
PDBTitle: crystal structure of dihydroxynapthoic acid synthetase (gk2873) from2 geobacillus kaustophilus hta426
|
| 70 | c3h0uB_ |
|
not modelled |
98.9 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:putative enoyl-coa hydratase;
PDBTitle: crystal structure of a putative enoyl-coa hydratase from2 streptomyces avermitilis
|
| 71 | d1ef8a_ |
|
not modelled |
98.9 |
15 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 72 | c3ot6A_ |
|
not modelled |
98.9 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:enoyl-coa hydratase/isomerase family protein;
PDBTitle: crystal structure of an enoyl-coa hydratase/isomerase family protein2 from psudomonas syringae
|
| 73 | d1rjma_ |
|
not modelled |
98.9 |
19 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 74 | d1q52a_ |
|
not modelled |
98.9 |
16 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 75 | c3h81A_ |
|
not modelled |
98.9 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase echa8;
PDBTitle: crystal structure of enoyl-coa hydratase from mycobacterium2 tuberculosis
|
| 76 | c3qmjA_ |
|
not modelled |
98.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase, echa8_6;
PDBTitle: crystal structure of enoyl-coa hydratase echa8_6 from mycobacterium2 marinum
|
| 77 | c3sllC_ |
|
not modelled |
98.9 |
15 |
PDB header:isomerase
Chain: C: PDB Molecule:probable enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of a probable enoyl-coa hydratase/isomerase from2 mycobacterium abscessus
|
| 78 | c3hp0B_ |
|
not modelled |
98.9 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:putative polyketide biosynthesis enoyl-coa
PDBTitle: crystal structure of a putative polyketide biosynthesis2 enoyl-coa hydratase (pksh) from bacillus subtilis
|
| 79 | c1rjnC_ |
|
not modelled |
98.8 |
18 |
PDB header:lyase
Chain: C: PDB Molecule:menb;
PDBTitle: the crystal structure of menb (rv0548c) from mycobacterium2 tuberculosis in complex with the coa portion of naphthoyl coa
|
| 80 | c3hinA_ |
|
not modelled |
98.8 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:putative 3-hydroxybutyryl-coa dehydratase;
PDBTitle: crystal structure of putative enoyl-coa hydratase from2 rhodopseudomonas palustris cga009
|
| 81 | d2f6ia1 |
|
not modelled |
98.8 |
17 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Clp protease, ClpP subunit |
| 82 | c3ju1A_ |
|
not modelled |
98.8 |
15 |
PDB header:lyase, isomerase
Chain: A: PDB Molecule:enoyl-coa hydratase/isomerase family protein;
PDBTitle: crystal structure of enoyl-coa hydratase/isomerase family protein
|
| 83 | c3h02F_ |
|
not modelled |
98.8 |
17 |
PDB header:lyase
Chain: F: PDB Molecule:naphthoate synthase;
PDBTitle: 2.15 angstrom resolution crystal structure of naphthoate synthase from2 salmonella typhimurium.
|
| 84 | c3p85A_ |
|
not modelled |
98.8 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure enoyl-coa hydratase from mycobacterium avium
|
| 85 | c3gkbA_ |
|
not modelled |
98.8 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative enoyl-coa hydratase;
PDBTitle: crystal structure of a putative enoyl-coa hydratase from streptomyces2 avermitilis
|
| 86 | d1mj3a_ |
|
not modelled |
98.8 |
15 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 87 | c3bptA_ |
|
not modelled |
98.8 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:3-hydroxyisobutyryl-coa hydrolase;
PDBTitle: crystal structure of human beta-hydroxyisobutyryl-coa hydrolase in2 complex with quercetin
|
| 88 | d1wdka4 |
|
not modelled |
98.8 |
14 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 89 | d1y7oa1 |
|
not modelled |
98.8 |
24 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Clp protease, ClpP subunit |
| 90 | c2j5iF_ |
|
not modelled |
98.8 |
17 |
PDB header:lyase
Chain: F: PDB Molecule:p-hydroxycinnamoyl coa hydratase/lyase;
PDBTitle: crystal structure of hydroxycinnamoyl-coa hydratase-lyase
|
| 91 | c2f6iG_ |
|
not modelled |
98.8 |
18 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp-dependent clp protease, putative;
PDBTitle: crystal structure of the clpp protease catalytic domain from2 plasmodium falciparum
|
| 92 | d2cbya1 |
|
not modelled |
98.8 |
18 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Clp protease, ClpP subunit |
| 93 | c2hw5F_ |
|
not modelled |
98.8 |
16 |
PDB header:lyase
Chain: F: PDB Molecule:enoyl-coa hydratase;
PDBTitle: the crystal structure of human enoyl-coenzyme a (coa) hydratase short2 chain 1, echs1
|
| 94 | c3q7hM_ |
|
not modelled |
98.8 |
22 |
PDB header:hydrolase
Chain: M: PDB Molecule:atp-dependent clp protease proteolytic subunit;
PDBTitle: structure of the clpp subunit of the atp-dependent clp protease from2 coxiella burnetii
|
| 95 | d1szoa_ |
|
not modelled |
98.8 |
10 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
| 96 | c3he2C_ |
|
not modelled |
98.8 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:enoyl-coa hydratase echa6;
PDBTitle: crystal structure of enoyl-coa hydratase from mycobacterium2 tuberculosis
|
| 97 | d1yg6a1 |
|
not modelled |
98.8 |
25 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Clp protease, ClpP subunit |
| 98 | c3qxiA_ |
|
not modelled |
98.8 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase echa1;
PDBTitle: crystal structure of enoyl-coa hydratase echa1 from mycobacterium2 marinum
|
| 99 | c1y7oE_ |
|
not modelled |
98.7 |
24 |
PDB header:hydrolase
Chain: E: PDB Molecule:atp-dependent clp protease proteolytic subunit;
PDBTitle: the structure of streptococcus pneumoniae a153p clpp
|
| 100 | c3qxzA_ |
|
not modelled |
98.7 |
18 |
PDB header:lyase,isomerase
Chain: A: PDB Molecule:enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of a probable enoyl-coa hydratase/isomerase from2 mycobacterium abscessus
|
| 101 | c2cbyG_ |
|
not modelled |
98.7 |
19 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp-dependent clp protease proteolytic subunit 1;
PDBTitle: crystal structure of the atp-dependent clp protease2 proteolytic subunit 1 (clpp1) from mycobacterium3 tuberculosis
|
| 102 | c2j5gL_ |
|
not modelled |
98.7 |
16 |
PDB header:hydrolase
Chain: L: PDB Molecule:alr4455 protein;
PDBTitle: the native structure of a beta-diketone hydrolase from the2 cyanobacterium anabaena sp. pcc 7120
|
| 103 | c3qkaB_ |
|
not modelled |
98.7 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:enoyl-coa hydratase, echa5;
PDBTitle: crystal structure of enoyl-coa hydratase echa5 from mycobacterium2 marinum
|
| 104 | c1tg6G_ |
|
not modelled |
98.7 |
20 |
PDB header:hydrolase
Chain: G: PDB Molecule:putative atp-dependent clp protease proteolytic subunit;
PDBTitle: crystallography and mutagenesis point to an essential role for the n-2 terminus of human mitochondrial clpp
|
| 105 | c3bezC_ |
|
not modelled |
98.7 |
21 |
PDB header:hydrolase
Chain: C: PDB Molecule:protease 4;
PDBTitle: crystal structure of escherichia coli signal peptide peptidase (sppa),2 semet crystals
|
| 106 | c3kthD_ |
|
not modelled |
98.7 |
23 |
PDB header:hydrolase
Chain: D: PDB Molecule:atp-dependent clp protease proteolytic subunit;
PDBTitle: structure of clpp from bacillus subtilis in orthorombic crystal form
|
| 107 | c3swxB_ |
|
not modelled |
98.7 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:probable enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of a probable enoyl-coa hydratase/isomerase from2 mycobacterium abscessus
|
| 108 | c3m6nA_ |
|
not modelled |
98.7 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:rpff protein;
PDBTitle: crystal structure of rpff
|
| 109 | c3p2lD_ |
|
not modelled |
98.7 |
24 |
PDB header:hydrolase
Chain: D: PDB Molecule:atp-dependent clp protease proteolytic subunit;
PDBTitle: crystal structure of atp-dependent clp protease subunit p from2 francisella tularensis
|
| 110 | c2d3tB_ |
|
not modelled |
98.7 |
13 |
PDB header:lyase, oxidoreductase/transferase
Chain: B: PDB Molecule:fatty oxidation complex alpha subunit;
PDBTitle: fatty acid beta-oxidation multienzyme complex from2 pseudomonas fragi, form v
|
| 111 | c3laoA_ |
|
not modelled |
98.7 |
15 |
PDB header:lyase, isomerase
Chain: A: PDB Molecule:enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of enoyl-coa hydratase from pseudomonas2 aeruginosa pa01
|
| 112 | d1tg6a1 |
|
not modelled |
98.7 |
20 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Clp protease, ClpP subunit |
| 113 | c3r0oA_ |
|
not modelled |
98.7 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:carnitinyl-coa dehydratase;
PDBTitle: crystal structure of carnitinyl-coa hydratase from mycobacterium avium
|
| 114 | c3trrA_ |
|
not modelled |
98.7 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:probable enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of a probable enoyl-coa hydratase/isomerase from2 mycobacterium abscessus
|
| 115 | c2x58B_ |
|
not modelled |
98.6 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peroxisomal bifunctional enzyme;
PDBTitle: the crystal structure of mfe1 liganded with coa
|
| 116 | c2deoA_ |
|
not modelled |
98.6 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:441aa long hypothetical nfed protein;
PDBTitle: 1510-n membrane protease specific for a stomatin homolog from2 pyrococcus horikoshii
|
| 117 | c3qreA_ |
|
not modelled |
98.6 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase, echa12_1;
PDBTitle: crystal structure of an enoyl-coa hydratase echa12_1 from2 mycobacterium marinum
|
| 118 | c2wtbA_ |
|
not modelled |
98.5 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fatty acid multifunctional protein (atmfp2);
PDBTitle: arabidopsis thaliana multifuctional protein, mfp2
|
| 119 | c2w3pB_ |
|
not modelled |
98.3 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:benzoyl-coa-dihydrodiol lyase;
PDBTitle: boxc crystal structure
|
| 120 | c2pg8C_ |
|
not modelled |
98.1 |
25 |
PDB header:ligand binding protein
Chain: C: PDB Molecule:dpgc;
PDBTitle: crystal structure of r254k mutanat of dpgc with bound substrate analog
|