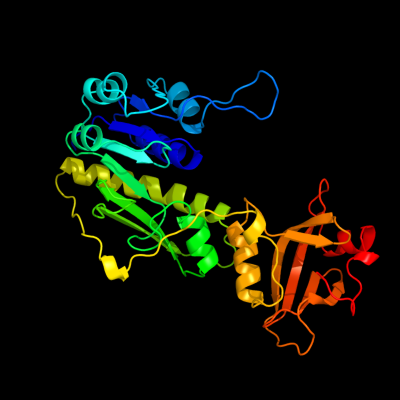
| 1 | c1fmtA_
|
|
 |
100.0 |
100 |
PDB header:formyltransferase
Chain: A: PDB Molecule:methionyl-trna fmet formyltransferase;
PDBTitle: methionyl-trnafmet formyltransferase from escherichia coli
|
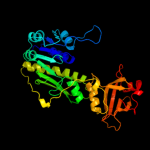
| 2 | c3rfoA_
|
|
 |
100.0 |
40 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: crystal structure of methyionyl-trna formyltransferase from bacillus2 anthracis
|
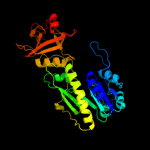
| 3 | c3q0iA_
|
|
 |
100.0 |
62 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: methionyl-trna formyltransferase from vibrio cholerae
|
| 4 | c3tqqA_
|
|
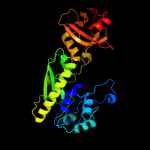 |
100.0 |
51 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: structure of the methionyl-trna formyltransferase (fmt) from coxiella2 burnetii
|
| 5 | c1z7eC_
|
|
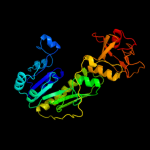 |
100.0 |
29 |
PDB header:hydrolase
Chain: C: PDB Molecule:protein arna;
PDBTitle: crystal structure of full length arna
|
| 6 | c1yrwA_
|
|
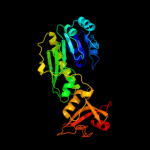 |
100.0 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:protein arna;
PDBTitle: crystal structure of e.coli arna transformylase domain
|
| 7 | c1s3iA_
|
|
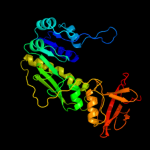 |
100.0 |
33 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:10-formyltetrahydrofolate dehydrogenase;
PDBTitle: crystal structure of the n terminal hydrolase domain of 10-2 formyltetrahydrofolate dehydrogenase
|
| 8 | d1fmta2
|
|
 |
100.0 |
100 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 9 | d2blna2
|
|
 |
100.0 |
30 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 10 | d2bw0a2
|
|
 |
100.0 |
36 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 11 | d1s3ia2
|
|
 |
100.0 |
39 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 12 | c1zghA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: methionyl-trna formyltransferase from clostridium thermocellum
|
| 13 | c3kcqA_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 anaplasma phagocytophilum
|
| 14 | d1jkxa_
|
|
 |
100.0 |
20 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 15 | c3dcjA_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:probable 5'-phosphoribosylglycinamide
PDBTitle: crystal structure of glycinamide formyltransferase (purn)2 from mycobacterium tuberculosis in complex with 5-methyl-5,3 6,7,8-tetrahydrofolic acid derivative
|
| 16 | c2ywrA_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of gar transformylase from aquifex2 aeolicus
|
| 17 | c3tqrA_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: structure of the phosphoribosylglycinamide formyltransferase (purn) in2 complex with ches from coxiella burnetii
|
| 18 | c3p9xB_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 bacillus halodurans
|
| 19 | d1meoa_
|
|
 |
100.0 |
23 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 20 | c3n0vD_
|
|
 |
100.0 |
20 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
| 21 | c3o1lB_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
|
| 22 | c3louB_ |
|
not modelled |
100.0 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
|
| 23 | c3obiC_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: C: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
|
| 24 | c3nrbD_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
|
| 25 | d1zgha2 |
|
not modelled |
100.0 |
19 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 26 | d1fmta1 |
|
not modelled |
100.0 |
100 |
Fold:FMT C-terminal domain-like
Superfamily:FMT C-terminal domain-like
Family:Post formyltransferase domain |
| 27 | d2blna1 |
|
not modelled |
99.9 |
28 |
Fold:FMT C-terminal domain-like
Superfamily:FMT C-terminal domain-like
Family:Post formyltransferase domain |
| 28 | d1s3ia1 |
|
not modelled |
99.9 |
25 |
Fold:FMT C-terminal domain-like
Superfamily:FMT C-terminal domain-like
Family:Post formyltransferase domain |
| 29 | d2bw0a1 |
|
not modelled |
99.9 |
20 |
Fold:FMT C-terminal domain-like
Superfamily:FMT C-terminal domain-like
Family:Post formyltransferase domain |
| 30 | c3rbvA_ |
|
not modelled |
96.5 |
26 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sugar 3-ketoreductase;
PDBTitle: crystal structure of kijd10, a 3-ketoreductase from actinomadura2 kijaniata incomplex with nadp
|
| 31 | c3ceaA_ |
|
not modelled |
96.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
| 32 | c3db2C_ |
|
not modelled |
96.4 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative nadph-dependent oxidoreductase;
PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
|
| 33 | d1vlva2 |
|
not modelled |
96.4 |
23 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 34 | c3euwB_ |
|
not modelled |
96.3 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:myo-inositol dehydrogenase;
PDBTitle: crystal structure of a myo-inositol dehydrogenase from corynebacterium2 glutamicum atcc 13032
|
| 35 | c3e18A_ |
|
not modelled |
96.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of nad-binding protein from listeria innocua
|
| 36 | c3q2kB_ |
|
not modelled |
95.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of the wlba dehydrogenase from bordetella pertussis2 in complex with nadh and udp-glcnaca
|
| 37 | c3v5nA_ |
|
not modelled |
95.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: the crystal structure of oxidoreductase from sinorhizobium meliloti
|
| 38 | c3e9mC_ |
|
not modelled |
95.9 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
| 39 | c2p2gD_ |
|
not modelled |
95.9 |
25 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from mycobacterium2 tuberculosis (rv1656): orthorhombic form
|
| 40 | c3ec7C_ |
|
not modelled |
95.7 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative dehydrogenase;
PDBTitle: crystal structure of putative dehydrogenase from salmonella2 typhimurium lt2
|
| 41 | c3nt5B_ |
|
not modelled |
95.7 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inositol 2-dehydrogenase/d-chiro-inositol 3-dehydrogenase;
PDBTitle: crystal structure of myo-inositol dehydrogenase from bacillus subtilis2 with bound cofactor and product inosose
|
| 42 | c1zh8B_ |
|
not modelled |
95.6 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
|
| 43 | d1lssa_ |
|
not modelled |
95.6 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
| 44 | c3ezyB_ |
|
not modelled |
95.5 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of probable dehydrogenase tm_0414 from2 thermotoga maritima
|
| 45 | c2ho3D_ |
|
not modelled |
95.3 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase, gfo/idh/moca family from2 streptococcus pneumoniae
|
| 46 | d1pvva2 |
|
not modelled |
95.3 |
17 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 47 | d1a9xa4 |
|
not modelled |
95.3 |
18 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 48 | d1zh8a1 |
|
not modelled |
95.2 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 49 | c3m2tA_ |
|
not modelled |
95.2 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of dehydrogenase from chromobacterium2 violaceum
|
| 50 | c3ic5A_ |
|
not modelled |
95.2 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
| 51 | c3eywA_ |
|
not modelled |
95.1 |
25 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
| 52 | c2pzlB_ |
|
not modelled |
95.1 |
17 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme2 wbmg in complex with nad and udp
|
| 53 | c1ml4A_ |
|
not modelled |
95.1 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:aspartate transcarbamoylase;
PDBTitle: the pala-liganded aspartate transcarbamoylase catalytic subunit from2 pyrococcus abyssi
|
| 54 | d1ekxa2 |
|
not modelled |
95.0 |
19 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 55 | c2p5uC_ |
|
not modelled |
95.0 |
21 |
PDB header:isomerase
Chain: C: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of thermus thermophilus hb8 udp-glucose 4-2 epimerase complex with nad
|
| 56 | d1kewa_ |
|
not modelled |
95.0 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 57 | d1qh8b_ |
|
not modelled |
94.9 |
17 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 58 | d1ml4a2 |
|
not modelled |
94.9 |
25 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 59 | c2o48X_ |
|
not modelled |
94.9 |
26 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:dimeric dihydrodiol dehydrogenase;
PDBTitle: crystal structure of mammalian dimeric dihydrodiol dehydrogenase
|
| 60 | d2nu7a1 |
|
not modelled |
94.8 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 61 | c2pk3B_ |
|
not modelled |
94.8 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gdp-6-deoxy-d-lyxo-4-hexulose reductase;
PDBTitle: crystal structure of a gdp-4-keto-6-deoxy-d-mannose reductase
|
| 62 | d1zgha1 |
|
not modelled |
94.8 |
15 |
Fold:FMT C-terminal domain-like
Superfamily:FMT C-terminal domain-like
Family:Post formyltransferase domain |
| 63 | d1ek6a_ |
|
not modelled |
94.7 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 64 | c3l4bG_ |
|
not modelled |
94.7 |
18 |
PDB header:transport protein
Chain: G: PDB Molecule:trka k+ channel protien tm1088b;
PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
|
| 65 | c1ofgF_ |
|
not modelled |
94.6 |
14 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
| 66 | c1h6dL_ |
|
not modelled |
94.5 |
16 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
| 67 | c3dtyA_ |
|
not modelled |
94.5 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from pseudomonas2 syringae
|
| 68 | c3moiA_ |
|
not modelled |
94.5 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of the putative dehydrogenase from bordetella2 bronchiseptica rb50
|
| 69 | d1miob_ |
|
not modelled |
94.5 |
14 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 70 | c1vm6B_ |
|
not modelled |
94.5 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: crystal structure of dihydrodipicolinate reductase (tm1520) from2 thermotoga maritima at 2.27 a resolution
|
| 71 | d1m1nb_ |
|
not modelled |
94.3 |
21 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 72 | c3gfgB_ |
|
not modelled |
94.2 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uncharacterized oxidoreductase yvaa;
PDBTitle: structure of putative oxidoreductase yvaa from bacillus subtilis in2 triclinic form
|
| 73 | d1tuga1 |
|
not modelled |
94.1 |
23 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 74 | c1vlvA_ |
|
not modelled |
94.1 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase (tm1097) from2 thermotoga maritima at 2.25 a resolution
|
| 75 | c2hunB_ |
|
not modelled |
94.0 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:336aa long hypothetical dtdp-glucose 4,6-dehydratase;
PDBTitle: crystal structure of hypothetical protein ph0414 from pyrococcus2 horikoshii ot3
|
| 76 | c3grfA_ |
|
not modelled |
94.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: x-ray structure of ornithine transcarbamoylase from giardia2 lamblia
|
| 77 | c2q4eB_ |
|
not modelled |
94.0 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable oxidoreductase at4g09670;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at4g09670
|
| 78 | d2nvwa1 |
|
not modelled |
93.9 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 79 | c3uuwB_ |
|
not modelled |
93.8 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase with nad(p)-binding rossmann-fold
PDBTitle: 1.63 angstrom resolution crystal structure of dehydrogenase (mvim)2 from clostridium difficile.
|
| 80 | c1a1sA_ |
|
not modelled |
93.8 |
21 |
PDB header:transcarbamylase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: ornithine carbamoyltransferase from pyrococcus furiosus
|
| 81 | d1pg5a2 |
|
not modelled |
93.7 |
21 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 82 | c2ph5A_ |
|
not modelled |
93.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:homospermidine synthase;
PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
|
| 83 | c2nvwB_ |
|
not modelled |
93.5 |
24 |
PDB header:transcription
Chain: B: PDB Molecule:galactose/lactose metabolism regulatory protein
PDBTitle: crystal sctucture of transcriptional regulator gal80p from2 kluyveromymes lactis
|
| 84 | d1o6ca_ |
|
not modelled |
93.5 |
11 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDP-N-acetylglucosamine 2-epimerase |
| 85 | c2glxD_ |
|
not modelled |
93.5 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:1,5-anhydro-d-fructose reductase;
PDBTitle: crystal structure analysis of bacterial 1,5-af reductase
|
| 86 | c3fd8A_ |
|
not modelled |
93.5 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
| 87 | c1drwA_ |
|
not modelled |
93.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: escherichia coli dhpr/nhdh complex
|
| 88 | c3sc6F_ |
|
not modelled |
93.4 |
22 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: 2.65 angstrom resolution crystal structure of dtdp-4-dehydrorhamnose2 reductase (rfbd) from bacillus anthracis str. ames in complex with3 nadp
|
| 89 | c2ixaA_ |
|
not modelled |
93.3 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-n-acetylgalactosaminidase;
PDBTitle: a-zyme, n-acetylgalactosaminidase
|
| 90 | c1kjjA_ |
|
not modelled |
93.3 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase 2;
PDBTitle: crystal structure of glycniamide ribonucleotide2 transformylase in complex with mg-atp-gamma-s
|
| 91 | c1pg5A_ |
|
not modelled |
93.2 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:aspartate carbamoyltransferase;
PDBTitle: crystal structure of the unligated (t-state) aspartate2 transcarbamoylase from the extremely thermophilic archaeon sulfolobus3 acidocaldarius
|
| 92 | c3tpfF_ |
|
not modelled |
93.2 |
16 |
PDB header:transferase
Chain: F: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of anabolic ornithine carbamoyltransferase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 93 | c3kuxA_ |
|
not modelled |
93.2 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: structure of the ypo2259 putative oxidoreductase from yersinia pestis
|
| 94 | c3evnA_ |
|
not modelled |
93.1 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of putative oxidoreductase from streptococcus2 agalactiae 2603v/r
|
| 95 | c2ys6A_ |
|
not modelled |
93.1 |
12 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylglycinamide synthetase;
PDBTitle: crystal structure of gar synthetase from geobacillus kaustophilus
|
| 96 | c3dapB_ |
|
not modelled |
93.1 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:diaminopimelic acid dehydrogenase;
PDBTitle: c. glutamicum dap dehydrogenase in complex with nadp+ and2 the inhibitor 5s-isoxazoline
|
| 97 | d1ru8a_ |
|
not modelled |
92.9 |
9 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 98 | d1ydwa1 |
|
not modelled |
92.9 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 99 | c3ot5D_ |
|
not modelled |
92.9 |
10 |
PDB header:isomerase
Chain: D: PDB Molecule:udp-n-acetylglucosamine 2-epimerase;
PDBTitle: 2.2 angstrom resolution crystal structure of putative udp-n-2 acetylglucosamine 2-epimerase from listeria monocytogenes
|
| 100 | c2axqA_ |
|
not modelled |
92.9 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
|
| 101 | c2qx7A_ |
|
not modelled |
92.8 |
16 |
PDB header:plant protein
Chain: A: PDB Molecule:eugenol synthase 1;
PDBTitle: structure of eugenol synthase from ocimum basilicum
|
| 102 | d1bxka_ |
|
not modelled |
92.8 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 103 | c3enkB_ |
|
not modelled |
92.7 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: 1.9a crystal structure of udp-glucose 4-epimerase from2 burkholderia pseudomallei
|
| 104 | c2xdqA_ |
|
not modelled |
92.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:light-independent protochlorophyllide reductase subunit n;
PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-2 chlb)2 complex
|
| 105 | c1i36A_ |
|
not modelled |
92.4 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein mth1747;
PDBTitle: structure of conserved protein mth1747 of unknown function2 reveals structural similarity with 3-hydroxyacid3 dehydrogenases
|
| 106 | c2rgwD_ |
|
not modelled |
92.4 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:aspartate carbamoyltransferase;
PDBTitle: catalytic subunit of m. jannaschii aspartate2 transcarbamoylase
|
| 107 | c2dzdB_ |
|
not modelled |
92.4 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of the biotin carboxylase domain of2 pyruvate carboxylase
|
| 108 | d1f06a1 |
|
not modelled |
92.4 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 109 | c3nklA_ |
|
not modelled |
92.2 |
10 |
PDB header:oxidoreductase/lyase
Chain: A: PDB Molecule:udp-d-quinovosamine 4-dehydrogenase;
PDBTitle: crystal structure of udp-d-quinovosamine 4-dehydrogenase from vibrio2 fischeri
|
| 110 | c3gd5D_ |
|
not modelled |
92.2 |
19 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from gloeobacter2 violaceus
|
| 111 | d1vl0a_ |
|
not modelled |
92.2 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 112 | c1xeaD_ |
|
not modelled |
92.1 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of a gfo/idh/moca family oxidoreductase2 from vibrio cholerae
|
| 113 | d1qh8a_ |
|
not modelled |
92.1 |
11 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 114 | c2yy7B_ |
|
not modelled |
92.1 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-threonine dehydrogenase;
PDBTitle: crystal structure of thermolabile l-threonine dehydrogenase from2 flavobacterium frigidimaris kuc-1
|
| 115 | d1udca_ |
|
not modelled |
92.1 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 116 | c2q1uA_ |
|
not modelled |
92.1 |
16 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmf in2 complex with nad+ and udp
|
| 117 | c2ggsB_ |
|
not modelled |
92.1 |
8 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:273aa long hypothetical dtdp-4-dehydrorhamnose
PDBTitle: crystal structure of hypothetical dtdp-4-dehydrorhamnose2 reductase from sulfolobus tokodaii
|
| 118 | c1fvoB_ |
|
not modelled |
91.9 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:ornithine transcarbamylase;
PDBTitle: crystal structure of human ornithine transcarbamylase complexed with2 carbamoyl phosphate
|
| 119 | d1gsoa2 |
|
not modelled |
91.9 |
18 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 120 | c2p2sA_ |
|
not modelled |
91.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase (yp_050235.1) from2 erwinia carotovora atroseptica scri1043 at 1.25 a resolution
|