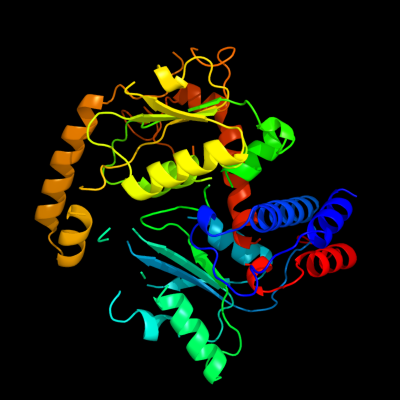
| 1 | c3q98A_
|
|
 |
100.0 |
96 |
PDB header:transferase
Chain: A: PDB Molecule:transcarbamylase;
PDBTitle: structure of ygew encoded protein from e. coli
|
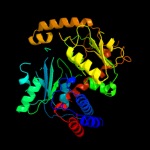
| 2 | c2otcA_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: ornithine transcarbamoylase complexed with n-2 (phosphonacetyl)-l-ornithine
|
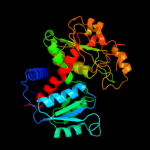
| 3 | c3updA_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: 2.9 angstrom crystal structure of ornithine carbamoyltransferase2 (argf) from vibrio vulnificus
|
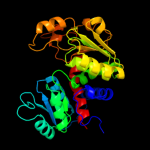
| 4 | c2w37A_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase, catabolic;
PDBTitle: crystal structure of the hexameric catabolic ornithine2 transcarbamylase from lactobacillus hilgardii
|
| 5 | c1ortD_
|
|
 |
100.0 |
26 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine transcarbamoylase;
PDBTitle: ornithine transcarbamoylase from pseudomonas aeruginosa
|
| 6 | c1fvoB_
|
|
 |
100.0 |
26 |
PDB header:transferase
Chain: B: PDB Molecule:ornithine transcarbamylase;
PDBTitle: crystal structure of human ornithine transcarbamylase complexed with2 carbamoyl phosphate
|
| 7 | c1a1sA_
|
|
 |
100.0 |
27 |
PDB header:transcarbamylase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: ornithine carbamoyltransferase from pyrococcus furiosus
|
| 8 | c1vlvA_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase (tm1097) from2 thermotoga maritima at 2.25 a resolution
|
| 9 | c1ml4A_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:aspartate transcarbamoylase;
PDBTitle: the pala-liganded aspartate transcarbamoylase catalytic subunit from2 pyrococcus abyssi
|
| 10 | c1zq2A_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of n-acetyl-l-ornithine transcarbamylase2 complexed with cp
|
| 11 | d1tuga1
|
|
 |
100.0 |
22 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 12 | c3sdsA_
|
|
 |
100.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase, mitochondrial;
PDBTitle: crystal structure of a mitochondrial ornithine carbamoyltransferase2 from coccidioides immitis
|
| 13 | c2p2gD_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from mycobacterium2 tuberculosis (rv1656): orthorhombic form
|
| 14 | c3tpfF_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: F: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of anabolic ornithine carbamoyltransferase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 15 | c1js1Z_
|
|
 |
100.0 |
26 |
PDB header:transferase
Chain: Z: PDB Molecule:transcarbamylase;
PDBTitle: crystal structure of a new transcarbamylase from the2 anaerobic bacterium bacteroides fragilis at 2.0 a3 resolution
|
| 16 | c1pg5A_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:aspartate carbamoyltransferase;
PDBTitle: crystal structure of the unligated (t-state) aspartate2 transcarbamoylase from the extremely thermophilic archaeon sulfolobus3 acidocaldarius
|
| 17 | c2rgwD_
|
|
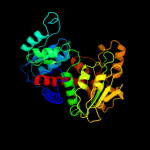 |
100.0 |
24 |
PDB header:transferase
Chain: D: PDB Molecule:aspartate carbamoyltransferase;
PDBTitle: catalytic subunit of m. jannaschii aspartate2 transcarbamoylase
|
| 18 | c2ef0A_
|
|
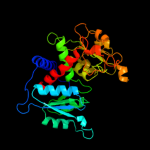 |
100.0 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from thermus2 thermophilus
|
| 19 | c3gd5D_
|
|
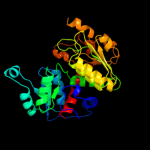 |
100.0 |
29 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from gloeobacter2 violaceus
|
| 20 | c3grfA_
|
|
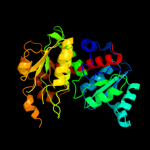 |
100.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: x-ray structure of ornithine transcarbamoylase from giardia2 lamblia
|
| 21 | c2at2B_ |
|
not modelled |
100.0 |
24 |
PDB header:
PDB COMPND:
|
| 22 | c3d6nB_ |
|
not modelled |
100.0 |
18 |
PDB header:hydrolase/transferase
Chain: B: PDB Molecule:aspartate carbamoyltransferase;
PDBTitle: crystal structure of aquifex dihydroorotase activated by aspartate2 transcarbamoylase
|
| 23 | c3lxmC_ |
|
not modelled |
100.0 |
24 |
PDB header:transferase
Chain: C: PDB Molecule:aspartate carbamoyltransferase;
PDBTitle: 2.00 angstrom resolution crystal structure of a catalytic2 subunit of an aspartate carbamoyltransferase (pyrb) from3 yersinia pestis co92
|
| 24 | d1ml4a1 |
|
not modelled |
100.0 |
25 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 25 | d1vlva1 |
|
not modelled |
100.0 |
26 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 26 | d1pvva1 |
|
not modelled |
100.0 |
28 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 27 | d1duvg1 |
|
not modelled |
100.0 |
30 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 28 | d1dxha1 |
|
not modelled |
100.0 |
27 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 29 | d1otha1 |
|
not modelled |
100.0 |
28 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 30 | d1ekxa1 |
|
not modelled |
100.0 |
31 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 31 | d1dxha2 |
|
not modelled |
100.0 |
24 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 32 | d2at2a1 |
|
not modelled |
100.0 |
27 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 33 | d1duvg2 |
|
not modelled |
100.0 |
21 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 34 | d1otha2 |
|
not modelled |
100.0 |
24 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 35 | d1pg5a1 |
|
not modelled |
100.0 |
22 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 36 | d1pvva2 |
|
not modelled |
100.0 |
28 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 37 | d1js1x1 |
|
not modelled |
100.0 |
23 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 38 | d1vlva2 |
|
not modelled |
100.0 |
34 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 39 | d1js1x2 |
|
not modelled |
100.0 |
30 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 40 | d1ml4a2 |
|
not modelled |
100.0 |
17 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 41 | d1ekxa2 |
|
not modelled |
100.0 |
15 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 42 | d1pg5a2 |
|
not modelled |
100.0 |
14 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 43 | d2at2a2 |
|
not modelled |
100.0 |
21 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 44 | d2atca2 |
|
not modelled |
100.0 |
16 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 45 | c3gvpB_ |
|
not modelled |
97.8 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:adenosylhomocysteinase 3;
PDBTitle: human sahh-like domain of human adenosylhomocysteinase 3
|
| 46 | c3l07B_ |
|
not modelled |
97.8 |
16 |
PDB header:oxidoreductase,hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate2 cyclohydrolase, putative bifunctional protein fold from francisella3 tularensis.
|
| 47 | c3d4oA_ |
|
not modelled |
97.7 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 48 | c4a26B_ |
|
not modelled |
97.7 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative c-1-tetrahydrofolate synthase, cytoplasmic;
PDBTitle: the crystal structure of leishmania major n5,n10-2 methylenetetrahydrofolate dehydrogenase/cyclohydrolase
|
| 49 | c3d64A_ |
|
not modelled |
97.7 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 burkholderia pseudomallei
|
| 50 | c1b0aA_ |
|
not modelled |
97.6 |
18 |
PDB header:oxidoreductase,hydrolase
Chain: A: PDB Molecule:protein (fold bifunctional protein);
PDBTitle: 5,10, methylene-tetrahydropholate2 dehydrogenase/cyclohydrolase from e coli.
|
| 51 | c1gpjA_ |
|
not modelled |
97.5 |
20 |
PDB header:reductase
Chain: A: PDB Molecule:glutamyl-trna reductase;
PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
|
| 52 | c3oneA_ |
|
not modelled |
97.5 |
25 |
PDB header:hydrolase/hydrolase substrate
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of lupinus luteus s-adenosyl-l-homocysteine2 hydrolase in complex with adenine
|
| 53 | c3p2oB_ |
|
not modelled |
97.5 |
16 |
PDB header:oxidoreductase, hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
| 54 | c3n58D_ |
|
not modelled |
97.5 |
25 |
PDB header:hydrolase
Chain: D: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from brucella2 melitensis in ternary complex with nad and adenosine, orthorhombic3 form
|
| 55 | c1d4fD_ |
|
not modelled |
97.4 |
20 |
PDB header:hydrolase
Chain: D: PDB Molecule:s-adenosylhomocysteine hydrolase;
PDBTitle: crystal structure of recombinant rat-liver d244e mutant s-2 adenosylhomocysteine hydrolase
|
| 56 | c3p2oA_ |
|
not modelled |
97.3 |
16 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
| 57 | c2rirA_ |
|
not modelled |
97.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
| 58 | d1a4ia1 |
|
not modelled |
97.2 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 59 | c4a5oB_ |
|
not modelled |
97.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of pseudomonas aeruginosa n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)
|
| 60 | d1gpja2 |
|
not modelled |
97.0 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 61 | c3nglA_ |
|
not modelled |
96.9 |
12 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of bifunctional 5,10-methylenetetrahydrofolate2 dehydrogenase / cyclohydrolase from thermoplasma acidophilum
|
| 62 | d1b0aa1 |
|
not modelled |
96.8 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 63 | c3n7uD_ |
|
not modelled |
96.3 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:formate dehydrogenase;
PDBTitle: nad-dependent formate dehydrogenase from higher-plant arabidopsis2 thaliana in complex with nad and azide
|
| 64 | d1pjca1 |
|
not modelled |
96.3 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 65 | c2eezG_ |
|
not modelled |
96.3 |
20 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:alanine dehydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase from themus thermophilus
|
| 66 | d2naca1 |
|
not modelled |
96.3 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 67 | c1v8bA_ |
|
not modelled |
96.3 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of a hydrolase
|
| 68 | c1vjtA_ |
|
not modelled |
96.2 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-glucosidase;
PDBTitle: crystal structure of alpha-glucosidase (tm0752) from thermotoga2 maritima at 2.50 a resolution
|
| 69 | d1l7da1 |
|
not modelled |
96.1 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 70 | c2nacA_ |
|
not modelled |
96.1 |
21 |
PDB header:oxidoreductase(aldehyde(d),nad+(a))
Chain: A: PDB Molecule:nad-dependent formate dehydrogenase;
PDBTitle: high resolution structures of holo and apo formate dehydrogenase
|
| 71 | c1pjcA_ |
|
not modelled |
96.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (l-alanine dehydrogenase);
PDBTitle: l-alanine dehydrogenase complexed with nad
|
| 72 | c2dbqA_ |
|
not modelled |
96.1 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glyoxylate reductase;
PDBTitle: crystal structure of glyoxylate reductase (ph0597) from pyrococcus2 horikoshii ot3, complexed with nadp (i41)
|
| 73 | c2c2xB_ |
|
not modelled |
96.1 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase-
PDBTitle: three dimensional structure of bifunctional2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase3 from mycobacterium tuberculosis
|
| 74 | d1mx3a1 |
|
not modelled |
96.1 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 75 | c2j6iC_ |
|
not modelled |
96.0 |
26 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:formate dehydrogenase;
PDBTitle: candida boidinii formate dehydrogenase (fdh) c-terminal2 mutant
|
| 76 | c1u8xX_ |
|
not modelled |
96.0 |
27 |
PDB header:hydrolase
Chain: X: PDB Molecule:maltose-6'-phosphate glucosidase;
PDBTitle: crystal structure of glva from bacillus subtilis, a metal-requiring,2 nad-dependent 6-phospho-alpha-glucosidase
|
| 77 | c1l7eC_ |
|
not modelled |
96.0 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nicotinamide nucleotide transhydrogenase,
PDBTitle: crystal structure of r. rubrum transhydrogenase domain i2 with bound nadh
|
| 78 | c3p2yA_ |
|
not modelled |
96.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alanine dehydrogenase/pyridine nucleotide transhydrogenase;
PDBTitle: crystal structure of alanine dehydrogenase/pyridine nucleotide2 transhydrogenase from mycobacterium smegmatis
|
| 79 | c3dhyC_ |
|
not modelled |
96.0 |
29 |
PDB header:hydrolase
Chain: C: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structures of mycobacterium tuberculosis s-adenosyl-l-2 homocysteine hydrolase in ternary complex with substrate and3 inhibitors
|
| 80 | c1gdhA_ |
|
not modelled |
95.8 |
13 |
PDB header:oxidoreductase(choh (d)-nad(p)+ (a))
Chain: A: PDB Molecule:d-glycerate dehydrogenase;
PDBTitle: crystal structure of a nad-dependent d-glycerate2 dehydrogenase at 2.4 angstroms resolution
|
| 81 | c1a4iB_ |
|
not modelled |
95.8 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase /
PDBTitle: human tetrahydrofolate dehydrogenase / cyclohydrolase
|
| 82 | c2omeA_ |
|
not modelled |
95.8 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:c-terminal-binding protein 2;
PDBTitle: crystal structure of human ctbp2 dehydrogenase complexed with nad(h)
|
| 83 | c3kboB_ |
|
not modelled |
95.7 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxylate/hydroxypyruvate reductase a;
PDBTitle: 2.14 angstrom crystal structure of putative oxidoreductase (ycdw) from2 salmonella typhimurium in complex with nadp
|
| 84 | d1li4a1 |
|
not modelled |
95.6 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 85 | d1gdha1 |
|
not modelled |
95.5 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 86 | c2gcgB_ |
|
not modelled |
95.5 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxylate reductase/hydroxypyruvate reductase;
PDBTitle: ternary crystal structure of human glyoxylate2 reductase/hydroxypyruvate reductase
|
| 87 | c1wwkA_ |
|
not modelled |
95.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of phosphoglycerate dehydrogenase from pyrococcus2 horikoshii ot3
|
| 88 | c1obbB_ |
|
not modelled |
95.4 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-glucosidase;
PDBTitle: alpha-glucosidase a, agla, from thermotoga maritima in2 complex with maltose and nad+
|
| 89 | d1pjqa1 |
|
not modelled |
95.3 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
| 90 | c2qm3A_ |
|
not modelled |
95.3 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:predicted methyltransferase;
PDBTitle: crystal structure of a predicted methyltransferase from pyrococcus2 furiosus
|
| 91 | c1j4aA_ |
|
not modelled |
95.3 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: insights into domain closure, substrate specificity and2 catalysis of d-lactate dehydrogenase from lactobacillus3 bulgaricus
|
| 92 | c3bazA_ |
|
not modelled |
95.3 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hydroxyphenylpyruvate reductase;
PDBTitle: structure of hydroxyphenylpyruvate reductase from coleus blumei in2 complex with nadp+
|
| 93 | d1vjta1 |
|
not modelled |
95.3 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 94 | c2cukC_ |
|
not modelled |
95.3 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glycerate dehydrogenase/glyoxylate reductase;
PDBTitle: crystal structure of tt0316 protein from thermus thermophilus hb8
|
| 95 | c3oj0A_ |
|
not modelled |
95.3 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamyl-trna reductase;
PDBTitle: crystal structure of glutamyl-trna reductase from thermoplasma2 volcanium (nucleotide binding domain)
|
| 96 | c2eklA_ |
|
not modelled |
95.2 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: structure of st1218 protein from sulfolobus tokodaii
|
| 97 | d1v8ba1 |
|
not modelled |
95.2 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 98 | c2g76A_ |
|
not modelled |
95.1 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of human 3-phosphoglycerate dehydrogenase
|
| 99 | d1pzga1 |
|
not modelled |
95.0 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 100 | c3gg2B_ |
|
not modelled |
95.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:sugar dehydrogenase, udp-glucose/gdp-mannose
PDBTitle: crystal structure of udp-glucose 6-dehydrogenase from2 porphyromonas gingivalis bound to product udp-glucuronate
|
| 101 | c2bruB_ |
|
not modelled |
94.9 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad(p) transhydrogenase subunit alpha;
PDBTitle: complex of the domain i and domain iii of escherichia coli2 transhydrogenase
|
| 102 | c3evtA_ |
|
not modelled |
94.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of phosphoglycerate dehydrogenase from2 lactobacillus plantarum
|
| 103 | c1ybaC_ |
|
not modelled |
94.9 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: the active form of phosphoglycerate dehydrogenase
|
| 104 | d1j4aa1 |
|
not modelled |
94.8 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 105 | c1m75B_ |
|
not modelled |
94.8 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-hydroxyacyl-coa dehydrogenase;
PDBTitle: crystal structure of the n208s mutant of l-3-hydroxyacyl-2 coa dehydrogenase in complex with nad and acetoacetyl-coa
|
| 106 | c3oetF_ |
|
not modelled |
94.8 |
29 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:erythronate-4-phosphate dehydrogenase;
PDBTitle: d-erythronate-4-phosphate dehydrogenase complexed with nad
|
| 107 | d2dlda1 |
|
not modelled |
94.8 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 108 | c3gg9C_ |
|
not modelled |
94.7 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:d-3-phosphoglycerate dehydrogenase oxidoreductase protein;
PDBTitle: crystal structure of putative d-3-phosphoglycerate dehydrogenase2 oxidoreductase from ralstonia solanacearum
|
| 109 | c3dfzB_ |
|
not modelled |
94.7 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:precorrin-2 dehydrogenase;
PDBTitle: sirc, precorrin-2 dehydrogenase
|
| 110 | c3k5pA_ |
|
not modelled |
94.6 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of amino acid-binding act: d-isomer specific 2-2 hydroxyacid dehydrogenase catalytic domain from brucella melitensis
|
| 111 | c1xdwA_ |
|
not modelled |
94.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad+-dependent (r)-2-hydroxyglutarate
PDBTitle: nad+-dependent (r)-2-hydroxyglutarate dehydrogenase from2 acidaminococcus fermentans
|
| 112 | c1np3B_ |
|
not modelled |
94.6 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: crystal structure of class i acetohydroxy acid isomeroreductase from2 pseudomonas aeruginosa
|
| 113 | d1x7da_ |
|
not modelled |
94.6 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Ornithine cyclodeaminase-like |
| 114 | c2pi1C_ |
|
not modelled |
94.5 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: crystal structure of d-lactate dehydrogenase from aquifex2 aeolicus complexed with nad and lactic acid
|
| 115 | c2hjrK_ |
|
not modelled |
94.5 |
16 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of cryptosporidium parvum malate2 dehydrogenase
|
| 116 | d1np3a2 |
|
not modelled |
94.4 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 117 | c3c7cB_ |
|
not modelled |
94.3 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:octopine dehydrogenase;
PDBTitle: a structural basis for substrate and stereo selectivity in2 octopine dehydrogenase (odh-nadh-l-arginine)
|
| 118 | d1qp8a1 |
|
not modelled |
94.3 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 119 | d1ygya1 |
|
not modelled |
94.3 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 120 | c3k96B_ |
|
not modelled |
94.1 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
|