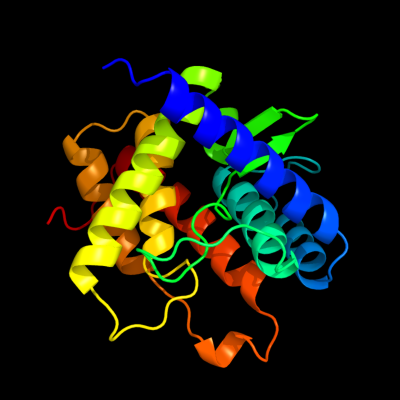
1 c2hp0A_
98.5
14
PDB header: isomeraseChain: A: PDB Molecule: ids-epimerase;PDBTitle: crystal structure of iminodisuccinate epimerase
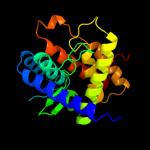
2 d1szqa_
98.0
14
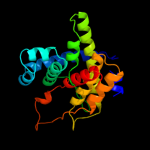
Fold: 2-methylcitrate dehydratase PrpDSuperfamily: 2-methylcitrate dehydratase PrpDFamily: 2-methylcitrate dehydratase PrpD3 c3u4gA_
56.6
18
PDB header: transferaseChain: A: PDB Molecule: namn:dmb phosphoribosyltransferase;PDBTitle: the structure of cobt from pyrococcus horikoshii
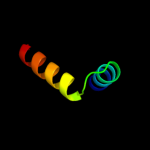
4 c3nepX_
53.0
18
PDB header: oxidoreductaseChain: X: PDB Molecule: malate dehydrogenase;PDBTitle: 1.55a resolution structure of malate dehydrogenase from salinibacter2 ruber
5 d1v59a2
47.3
12
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains6 c2o2jA_
38.7
27
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase beta chain;PDBTitle: mycobacterium tuberculosis tryptophan synthase beta chain2 dimer (apoform)
7 c2klzA_
28.3
57
PDB header: hydrolaseChain: A: PDB Molecule: ataxin-3;PDBTitle: solution structure of the tandem uim domain of ataxin-3 complexed with2 ubiquitin
8 c1x1qA_
23.0
23
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase beta chain;PDBTitle: crystal structure of tryptophan synthase beta chain from thermus2 thermophilus hb8
9 d1o58a_
20.3
23
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes10 c3l0zC_
19.5
20
PDB header: transferaseChain: C: PDB Molecule: putative nicotinate-nucleotide-dimethylbenzimidazolePDBTitle: crystal structure of a putative nicotinate-nucleotide-2 dimethylbenzimidazole phosphoribosyltransferase from3 methanocaldococcus jannaschii dsm 2661
11 d2hsga1
18.8
24
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator12 c1m75B_
18.5
39
PDB header: oxidoreductaseChain: B: PDB Molecule: 3-hydroxyacyl-coa dehydrogenase;PDBTitle: crystal structure of the n208s mutant of l-3-hydroxyacyl-2 coa dehydrogenase in complex with nad and acetoacetyl-coa
13 d1efaa1
16.7
18
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator14 d1e3ja2
15.6
35
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain15 c3e49A_
15.0
19
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein duf849 with a tim barrel fold;PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 with a tim barrel fold (bxe_c0966) from burkholderia xenovorans lb4003 at 1.75 a resolution
16 d1v71a1
14.8
19
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes17 c2hldH_
14.3
25
PDB header: hydrolaseChain: H: PDB Molecule: atp synthase delta chain, mitochondrial;PDBTitle: crystal structure of yeast mitochondrial f1-atpase
18 d1wfza_
13.7
13
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: NifU/IscU domain19 d1r4qa_
12.2
5
Fold: Ribosome inactivating proteins (RIP)Superfamily: Ribosome inactivating proteins (RIP)Family: Shiga toxin, A-chain20 d1fcja_
12.2
27
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes21 d1nvmb1
not modelled
12.1
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain22 c3doaA_
not modelled
11.6
17
PDB header: protein bindingChain: A: PDB Molecule: fibrinogen binding protein;PDBTitle: the crystal structure of the fibrinogen binding protein from2 staphylococcus aureus
23 c3rfuC_
not modelled
11.2
13
PDB header: hydrolase, membrane proteinChain: C: PDB Molecule: copper efflux atpase;PDBTitle: crystal structure of a copper-transporting pib-type atpase
24 c3co7C_
not modelled
10.9
29
PDB header: transcription/dnaChain: C: PDB Molecule: forkhead box protein o1;PDBTitle: crystal structure of foxo1 dbd bound to dbe2 dna
25 d1j0aa_
not modelled
10.7
18
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes26 c3a58A_
not modelled
10.6
20
PDB header: protein transport/exocytosisChain: A: PDB Molecule: exocyst complex component sec3;PDBTitle: crystal structure of sec3p - rho1p complex from2 saccharomyces cerevisiae
27 d3bpya1
not modelled
10.6
36
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain28 c3g73A_
not modelled
10.6
29
PDB header: transcription/dnaChain: A: PDB Molecule: forkhead box protein m1;PDBTitle: structure of the foxm1 dna binding
29 d1kq8a_
not modelled
10.5
43
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain30 c1kq8A_
not modelled
10.5
43
PDB header: transcriptionChain: A: PDB Molecule: hepatocyte nuclear factor 3 forkhead homolog 1;PDBTitle: solution structure of winged helix protein hfh-1
31 d2dx7a1
not modelled
10.5
23
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase32 d1tyza_
not modelled
10.5
11
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes33 c3c1zA_
not modelled
10.4
17
PDB header: dna binding proteinChain: A: PDB Molecule: dna integrity scanning protein disa;PDBTitle: structure of the ligand-free form of a bacterial dna damage2 sensor protein
34 d1ve1a1
not modelled
10.2
26
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes35 d2ldxa1
not modelled
10.2
27
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like36 d2bjca1
not modelled
10.2
18
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator37 d1j6ua3
not modelled
10.0
20
Fold: Ribokinase-likeSuperfamily: MurD-like peptide ligases, catalytic domainFamily: MurCDEF38 d2a07f1
not modelled
10.0
29
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain39 d2hfha_
not modelled
9.8
29
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain40 c1e17A_
not modelled
9.7
36
PDB header: dna binding domainChain: A: PDB Molecule: afx;PDBTitle: solution structure of the dna binding domain of the human2 forkhead transcription factor afx (foxo4)
41 d1ijwc_
not modelled
9.5
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain42 d1kn1b_
not modelled
9.4
23
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins43 d1pgja2
not modelled
9.2
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain44 c3imoC_
not modelled
9.1
33
PDB header: unknown functionChain: C: PDB Molecule: integron cassette protein;PDBTitle: structure from the mobile metagenome of vibrio cholerae.2 integron cassette protein vch_cass14
45 d2vnud2
not modelled
8.9
44
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like46 d1wkva1
not modelled
8.8
11
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes47 c2okvC_
not modelled
8.7
9
PDB header: hydrolaseChain: C: PDB Molecule: probable d-tyrosyl-trna(tyr) deacylase 1;PDBTitle: c-myc dna unwinding element binding protein
48 d1dxla2
not modelled
8.7
15
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains49 d1k1xa3
not modelled
8.7
15
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: 4-alpha-glucanotransferase, N-terminal domain50 c3mogA_
not modelled
8.6
39
PDB header: oxidoreductaseChain: A: PDB Molecule: probable 3-hydroxybutyryl-coa dehydrogenase;PDBTitle: crystal structure of 3-hydroxybutyryl-coa dehydrogenase from2 escherichia coli k12 substr. mg1655
51 c3sqgF_
not modelled
8.6
26
PDB header: transferaseChain: F: PDB Molecule: methyl-coenzyme m reductase, gamma subunit;PDBTitle: crystal structure of a methyl-coenzyme m reductase purified from black2 sea mats
52 d2bhsa1
not modelled
8.4
28
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes53 d1hcra_
not modelled
8.3
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain54 d1b74a2
not modelled
8.3
19
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase55 d2f1ka2
not modelled
8.2
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain56 c3dt5A_
not modelled
8.1
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein af_0924;PDBTitle: c_terminal domain of protein of unknown function af_0924 from2 archaeoglobus fulgidus.
57 d1j7ga_
not modelled
8.1
19
Fold: DTD-likeSuperfamily: DTD-likeFamily: DTD-like58 d1f2da_
not modelled
7.9
14
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes59 d2gv8a2
not modelled
7.9
21
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains60 d1qpza1
not modelled
7.7
14
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator61 c2yqfA_
not modelled
7.7
35
PDB header: protein bindingChain: A: PDB Molecule: ankyrin-1;PDBTitle: solution structure of the death domain of ankyrin-1
62 c3kg4A_
not modelled
7.7
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein from mannheimia2 succiniciproducens
63 d1gv4a2
not modelled
7.5
19
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains64 c2pqmA_
not modelled
7.4
27
PDB header: lyaseChain: A: PDB Molecule: cysteine synthase;PDBTitle: crystal structure of cysteine synthase (oass) from entamoeba2 histolytica at 1.86 a resolution
65 c1nvmB_
not modelled
7.4
24
PDB header: lyase/oxidoreductaseChain: B: PDB Molecule: acetaldehyde dehydrogenase (acylating);PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
66 d1iioa_
not modelled
7.3
28
Fold: EF Hand-likeSuperfamily: Hypothetical protein MTH865Family: Hypothetical protein MTH86567 d2c6ya1
not modelled
7.2
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain68 c2zskA_
not modelled
7.1
100
PDB header: unknown functionChain: A: PDB Molecule: 226aa long hypothetical aspartate racemase;PDBTitle: crystal structure of ph1733, an aspartate racemase2 homologue, from pyrococcus horikoshii ot3
69 c1i7mD_
not modelled
7.1
15
PDB header: lyaseChain: D: PDB Molecule: s-adenosylmethionine decarboxylase beta chain;PDBTitle: human s-adenosylmethionine decarboxylase with covalently bound2 pyruvoyl group and complexed with 4-amidinoindan-1-one-2'-3 amidinohydrazone
70 c3j08A_
not modelled
7.0
13
PDB header: hydrolase, metal transportChain: A: PDB Molecule: copper-exporting p-type atpase a;PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa
71 c2vmlA_
not modelled
7.0
20
PDB header: photosynthesisChain: A: PDB Molecule: phycocyanin alpha chain;PDBTitle: the monoclinic structure of phycocyanin from gloeobacter2 violaceus
72 d1dlja2
not modelled
6.9
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain73 d1jsca3
not modelled
6.9
17
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module74 c1dliA_
not modelled
6.9
25
PDB header: oxidoreductaseChain: A: PDB Molecule: udp-glucose dehydrogenase;PDBTitle: the first structure of udp-glucose dehydrogenase (udpgdh) reveals the2 catalytic residues necessary for the two-fold oxidation
75 d1d5va_
not modelled
6.9
29
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain76 c2jfzB_
not modelled
6.8
80
PDB header: isomeraseChain: B: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of helicobacter pylori glutamate racemase2 in complex with d-glutamate and an inhibitor
77 c1p8cD_
not modelled
6.7
28
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of tm1620 (apc4843) from thermotoga2 maritima
78 c3d3kD_
not modelled
6.6
14
PDB header: protein bindingChain: D: PDB Molecule: enhancer of mrna-decapping protein 3;PDBTitle: crystal structure of human edc3p
79 d7mdha1
not modelled
6.4
26
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like80 c2pwzG_
not modelled
6.4
22
PDB header: oxidoreductaseChain: G: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of the apo form of e.coli malate dehydrogenase
81 c2q2kA_
not modelled
6.4
24
PDB header: dna binding protein/dnaChain: A: PDB Molecule: hypothetical protein;PDBTitle: structure of nucleic-acid binding protein
82 d1dd3a1
not modelled
6.3
12
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain83 c2kdtA_
not modelled
6.2
21
PDB header: protein transportChain: A: PDB Molecule: neuroendocrine convertase 1;PDBTitle: pc1/3 dcsg sorting domain structure in dpc
84 d1allb_
not modelled
6.2
19
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins85 c1o7dA_
not modelled
6.1
12
PDB header: hydrolaseChain: A: PDB Molecule: lysosomal alpha-mannosidase;PDBTitle: the structure of the bovine lysosomal a-mannosidase2 suggests a novel mechanism for low ph activation
86 c3m9hB_
not modelled
6.1
44
PDB header: chaperoneChain: B: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain of the2 mycobacterium tuberculosis proteasomal atpase mpa
87 d1poia_
not modelled
6.0
22
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like88 d2nmla1
not modelled
6.0
8
Fold: ERH-likeSuperfamily: ERH-likeFamily: ERH-like89 d3bvua3
not modelled
6.0
12
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: alpha-mannosidase90 d1v8za1
not modelled
6.0
21
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes91 d1neka1
not modelled
6.0
9
Fold: Spectrin repeat-likeSuperfamily: Succinate dehydrogenase/fumarate reductase flavoprotein C-terminal domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein C-terminal domain92 c3jvfC_
not modelled
6.0
44
PDB header: signaling protein / cytokineChain: C: PDB Molecule: interleukin-17 receptor a;PDBTitle: crystal structure of an interleukin-17 receptor complex
93 c2jnhA_
not modelled
5.9
33
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase cbl-b;PDBTitle: solution structure of the uba domain from cbl-b
94 c1p5jA_
not modelled
5.9
21
PDB header: lyaseChain: A: PDB Molecule: l-serine dehydratase;PDBTitle: crystal structure analysis of human serine dehydratase
95 d1p5ja_
not modelled
5.9
21
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes96 d1ez4a1
not modelled
5.9
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like97 c2jfnA_
not modelled
5.9
80
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of escherichia coli glutamate racemase2 in complex with l-glutamate and activator udp-murnac-ala
98 c1b74A_
not modelled
5.8
80
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: glutamate racemase from aquifex pyrophilus
99 c3a3jA_
not modelled
5.8
16
PDB header: hydrolaseChain: A: PDB Molecule: pbp5;PDBTitle: crystal structures of penicillin binding protein 5 from2 haemophilus influenzae