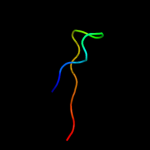
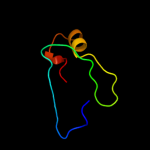
1 c3epsB_
100.0
99
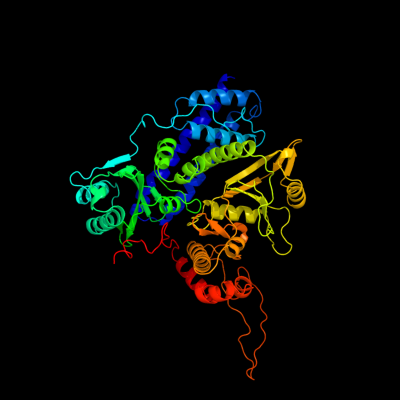
PDB header: transferase, hydrolaseChain: B: PDB Molecule: isocitrate dehydrogenase kinase/phosphatase;PDBTitle: the crystal structure of isocitrate dehydrogenase kinase/phosphatase2 from e. coli
2 c2an7A_
60.7
33
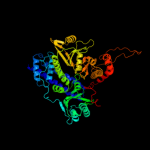
PDB header: dna binding proteinChain: A: PDB Molecule: protein pard;PDBTitle: solution structure of the bacterial antidote pard
3 d1o5ua_
49.4
22
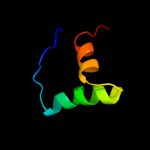
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Hypothetical protein TM11124 c3pcsB_
47.3
35
PDB header: protein transport/transferaseChain: B: PDB Molecule: espg;PDBTitle: structure of espg-pak2 autoinhibitory ialpha3 helix complex
5 c3es4B_
46.9
17
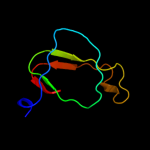
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein duf861 with a rmlc-like cupin fold;PDBTitle: crystal structure of protein of unknown function (duf861) with a rmlc-2 like cupin fold (17741406) from agrobacterium tumefaciens str. c583 (dupont) at 1.64 a resolution
6 c3myxA_
40.7
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pspto_0244;PDBTitle: crystal structure of a pspto_0244 (protein with unknown function which2 belongs to pfam duf861 family) from pseudomonas syringae pv. tomato3 str. dc3000 at 1.30 a resolution
7 c3rmiA_
39.2
16
PDB header: isomeraseChain: A: PDB Molecule: chorismate mutase protein;PDBTitle: crystal structure of chorismate mutase from bartonella henselae str.2 houston-1 in complex with malate
8 c3hm5A_
38.0
36
PDB header: transcriptionChain: A: PDB Molecule: dna methyltransferase 1-associated protein 1;PDBTitle: sant domain of human dna methyltransferase 1 associated2 protein 1
9 c3hzpA_
36.5
53
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ntf2-like protein of unknown function;PDBTitle: crystal structure of ntf2-like protein of unknown function mn2a_05052 from prochlorococcus marinus (yp_291699.1) from prochlorococcus sp.3 natl2a at 1.40 a resolution
10 c1nvpB_
35.0
23
PDB header: transcription/dnaChain: B: PDB Molecule: transcription initiation factor iia alpha chain;PDBTitle: human tfiia/tbp/dna complex
11 d1nvpb_
35.0
23
Fold: Transcription factor IIA (TFIIA), alpha-helical domainSuperfamily: Transcription factor IIA (TFIIA), alpha-helical domainFamily: Transcription factor IIA (TFIIA), alpha-helical domain12 d1nh2b_
34.3
22
Fold: Transcription factor IIA (TFIIA), alpha-helical domainSuperfamily: Transcription factor IIA (TFIIA), alpha-helical domainFamily: Transcription factor IIA (TFIIA), alpha-helical domain13 d1dwka2
32.7
19
Fold: Cyanase C-terminal domainSuperfamily: Cyanase C-terminal domainFamily: Cyanase C-terminal domain14 d2pyta1
30.1
45
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: EutQ-like15 d1oe4a_
29.8
17
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Single-strand selective monofunctional uracil-DNA glycosylase SMUG116 c3ff6D_
27.7
28
PDB header: ligaseChain: D: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: human acc2 ct domain with cp-640186
17 c2wk1A_
26.0
26
PDB header: transferaseChain: A: PDB Molecule: novp;PDBTitle: structure of the o-methyltransferase novp
18 c2b3fD_
26.0
14
PDB header: sugar binding proteinChain: D: PDB Molecule: glucose-binding protein;PDBTitle: thermus thermophilus glucose/galactose binding protein2 bound with galactose
19 d1zl8a1
25.2
29
Fold: L27 domainSuperfamily: L27 domainFamily: L27 domain20 c3fewX_
24.2
42
PDB header: immune systemChain: X: PDB Molecule: colicin s4;PDBTitle: structure and function of colicin s4, a colicin with a2 duplicated receptor binding domain
21 c3ic7A_
not modelled
22.8
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator of gntr family2 from bacteroides thetaiotaomicron
22 c2w7yA_
not modelled
22.7
22
PDB header: sugar-binding proteinChain: A: PDB Molecule: probable sugar abc transporter, sugar-bindingPDBTitle: structure of a streptococcus pneumoniae solute-binding2 protein in complex with the blood group a-trisaccharide.
23 c2og5A_
not modelled
22.3
14
PDB header: oxidoreductaseChain: A: PDB Molecule: putative oxygenase;PDBTitle: crystal structure of asparagine oxygenase (asno)
24 c3kpaB_
not modelled
22.2
26
PDB header: ligaseChain: B: PDB Molecule: probable ubiquitin fold modifier conjugating enzyme;PDBTitle: ubiquitin fold modifier conjugating enzyme from leishmania major2 (probable)
25 d1f3va_
not modelled
21.5
19
Fold: Ferredoxin-likeSuperfamily: TRADD, N-terminal domainFamily: TRADD, N-terminal domain26 c3ckxA_
not modelled
21.1
19
PDB header: transferaseChain: A: PDB Molecule: serine/threonine-protein kinase 24;PDBTitle: crystal structure of sterile 20-like kinase 3 (mst3, stk24)2 in complex with staurosporine
27 c3nquA_
not modelled
20.8
16
PDB header: dna binding proteinChain: A: PDB Molecule: histone h3-like centromeric protein a;PDBTitle: crystal structure of partially trypsinized (cenp-a/h4)2 heterotetramer
28 d2gtvx1
not modelled
20.6
10
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: monomeric chorismate mutase29 c1rm1C_
not modelled
20.3
23
PDB header: transcription/dnaChain: C: PDB Molecule: transcription initiation factor iia large chain;PDBTitle: structure of a yeast tfiia/tbp/tata-box dna complex
30 c2z8fB_
not modelled
20.0
19
PDB header: sugar binding proteinChain: B: PDB Molecule: galacto-n-biose/lacto-n-biose i transporter substrate-PDBTitle: the galacto-n-biose-/lacto-n-biose i-binding protein (gl-bp) of the2 abc transporter from bifidobacterium longum in complex with lacto-n-3 tetraose
31 c3i3vC_
not modelled
19.8
14
PDB header: transport proteinChain: C: PDB Molecule: probable secreted solute-binding lipoprotein;PDBTitle: crystal structure of probable secreted solute-binding2 lipoprotein from streptomyces coelicolor
32 c2l0rA_
not modelled
19.4
21
PDB header: hydrolase,toxinChain: A: PDB Molecule: lethal factor;PDBTitle: conformational dynamics of the anthrax lethal factor catalytic center
33 d1ppjw_
not modelled
18.8
7
Fold: Single transmembrane helixSuperfamily: Subunit X (non-heme 7 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: Subunit X (non-heme 7 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)34 c2hjvB_
not modelled
18.7
14
PDB header: hydrolaseChain: B: PDB Molecule: atp-dependent rna helicase dbpa;PDBTitle: structure of the second domain (residues 207-368) of the2 bacillus subtilis yxin protein
35 c3koxA_
not modelled
18.1
50
PDB header: metal binding proteinChain: A: PDB Molecule: d-ornithine aminomutase e component;PDBTitle: crystal structure of ornithine 4,5 aminomutase in complex with 2,4-2 diaminobutyrate (anaerobic)
36 c3lj4i_
not modelled
17.3
20
PDB header: viral proteinChain: I: PDB Molecule: portal protein;PDBTitle: bacteriophage p22 portal protein bound to middle tail factor gp4. this2 file contain the first biological assembly
37 d1p3ie_
not modelled
17.3
13
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones38 d1miua1
not modelled
17.2
33
Fold: BRCA2 helical domainSuperfamily: BRCA2 helical domainFamily: BRCA2 helical domain39 c2iv1J_
not modelled
16.7
19
PDB header: lyaseChain: J: PDB Molecule: cyanate hydratase;PDBTitle: site directed mutagenesis of key residues involved in the2 catalytic mechanism of cyanase
40 d1ckma2
not modelled
16.6
23
Fold: ATP-graspSuperfamily: DNA ligase/mRNA capping enzyme, catalytic domainFamily: mRNA capping enzyme41 d2ihoa2
not modelled
16.4
26
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: MOA C-terminal domain-like42 d2ff4a2
not modelled
16.2
5
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: BTAD-like43 c3bcwB_
not modelled
16.1
31
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf861 family protein with a rmlc-like cupin2 fold (bb1179) from bordetella bronchiseptica rb50 at 1.60 a3 resolution
44 d1pc2a_
not modelled
15.5
22
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)45 d1t5ia_
not modelled
15.3
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain46 c3oqhB_
not modelled
15.2
23
PDB header: ligaseChain: B: PDB Molecule: putative uncharacterized protein yvmc;PDBTitle: crystal structure of b. licheniformis cdps yvmc-blic
47 c2dnrA_
not modelled
15.2
35
PDB header: rna binding proteinChain: A: PDB Molecule: synaptojanin-1;PDBTitle: solution structure of rna binding domain in synaptojanin 1
48 d2h9ec1
not modelled
15.1
33
Fold: Serine protease inhibitorsSuperfamily: Serine protease inhibitorsFamily: ATI-like49 d1mvfd_
not modelled
14.5
21
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: Kis/PemI addiction antidote50 c3ls1A_
not modelled
14.3
15
PDB header: photosynthesisChain: A: PDB Molecule: sll1638 protein;PDBTitle: crystal structure of cyanobacterial psbq from synechocystis2 sp. pcc 6803 complexed with zn2+
51 c2ihoA_
not modelled
14.3
26
PDB header: sugar binding proteinChain: A: PDB Molecule: lectin;PDBTitle: crystal structure of moa, a lectin from the mushroom marasmius oreades2 in complex with the trisaccharide gal(1,3)gal(1,4)glcnac
52 c2a8vA_
not modelled
14.1
24
PDB header: protein/rnaChain: A: PDB Molecule: rna binding domain of rho transcriptionPDBTitle: rho transcription termination factor/rna complex
53 d2e29a1
not modelled
13.8
38
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: GUCT domain54 c2vwiC_
not modelled
13.6
14
PDB header: transferaseChain: C: PDB Molecule: serine/threonine-protein kinase osr1;PDBTitle: structure of the osr1 kinase, a hypertension drug target
55 d1a62a2
not modelled
13.4
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like56 d2enda_
not modelled
13.2
15
Fold: T4 endonuclease VSuperfamily: T4 endonuclease VFamily: T4 endonuclease V57 c2qbvA_
not modelled
13.1
20
PDB header: isomeraseChain: A: PDB Molecule: chorismate mutase;PDBTitle: crystal structure of intracellular chorismate mutase from2 mycobacterium tuberculosis
58 c3dkqB_
not modelled
12.9
22
PDB header: oxidoreductaseChain: B: PDB Molecule: pkhd-type hydroxylase sbal_3634;PDBTitle: crystal structure of putative oxygenase (yp_001051978.1) from2 shewanella baltica os155 at 2.26 a resolution
59 c2q6tB_
not modelled
12.9
10
PDB header: hydrolaseChain: B: PDB Molecule: dnab replication fork helicase;PDBTitle: crystal structure of the thermus aquaticus dnab monomer
60 d1k28d2
not modelled
12.9
36
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like61 d1ht6a2
not modelled
12.8
27
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain62 c1yt8A_
not modelled
12.6
35
PDB header: transferaseChain: A: PDB Molecule: thiosulfate sulfurtransferase;PDBTitle: crystal structure of thiosulfate sulfurtransferase from pseudomonas2 aeruginosa
63 c3fgrA_
not modelled
12.6
22
PDB header: hydrolaseChain: A: PDB Molecule: putative phospholipase b-like 2 28 kda form;PDBTitle: two chain form of the 66.3 kda protein at 1.8 angstroem
64 d1z84a1
not modelled
12.5
26
Fold: HIT-likeSuperfamily: HIT-likeFamily: Hexose-1-phosphate uridylyltransferase65 c3qufB_
not modelled
12.4
28
PDB header: transport proteinChain: B: PDB Molecule: extracellular solute-binding protein, family 1;PDBTitle: the structure of a family 1 extracellular solute-binding protein from2 bifidobacterium longum subsp. infantis
66 c3iz5G_
not modelled
12.3
38
PDB header: ribosomeChain: G: PDB Molecule: 60s ribosomal protein l6 (l6e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
67 c1tqmA_
not modelled
12.3
13
PDB header: ribosomeChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of a. fulgidus rio2 serine protein kinase bound to2 amppnp
68 d2fp1a1
not modelled
12.0
8
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Secreted chorismate mutase-like69 d2d8da1
not modelled
11.9
12
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Dimeric chorismate mutase70 d1t56a2
not modelled
11.9
10
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain71 d3dwya1
not modelled
11.8
9
Fold: Bromodomain-likeSuperfamily: BromodomainFamily: Bromodomain72 c2gbbA_
not modelled
11.7
8
PDB header: isomeraseChain: A: PDB Molecule: putative chorismate mutase;PDBTitle: crystal structure of secreted chorismate mutase from2 yersinia pestis
73 d1ecma_
not modelled
11.5
9
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Dimeric chorismate mutase74 c2xvtC_
not modelled
11.5
15
PDB header: membrane proteinChain: C: PDB Molecule: receptor activity-modifying protein 2;PDBTitle: structure of the extracellular domain of human ramp2
75 d2ok5a4
not modelled
11.4
27
Fold: Complement control module/SCR domainSuperfamily: Complement control module/SCR domainFamily: Complement control module/SCR domain76 c3olhA_
not modelled
11.3
22
PDB header: transferaseChain: A: PDB Molecule: 3-mercaptopyruvate sulfurtransferase;PDBTitle: human 3-mercaptopyruvate sulfurtransferase
77 d1j03a_
not modelled
11.1
23
Fold: Cytochrome b5-like heme/steroid binding domainSuperfamily: Cytochrome b5-like heme/steroid binding domainFamily: Steroid-binding domain78 d1nz6a_
not modelled
11.1
22
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain79 c2i58B_
not modelled
11.0
25
PDB header: sugar binding proteinChain: B: PDB Molecule: sugar abc transporter, sugar-binding protein;PDBTitle: crystal structure of rafe from streptococcus pneumoniae complexed with2 raffinose
80 d1cz5a1
not modelled
10.9
12
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Cdc48 N-terminal domain-like81 c1x58A_
not modelled
10.9
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein 4930532d21rik;PDBTitle: solution structures of the myb-like dna binding domain of2 4930532d21rik protein
82 d1y74a1
not modelled
10.8
19
Fold: L27 domainSuperfamily: L27 domainFamily: L27 domain83 c3r9jD_
not modelled
10.7
12
PDB header: cell cycle,hydrolase/cell cycleChain: D: PDB Molecule: cell division topological specificity factor;PDBTitle: 4.3a resolution structure of a mind-mine(i24n) protein complex
84 d1njha_
not modelled
10.7
20
Fold: Hypothetical protein YojFSuperfamily: Hypothetical protein YojFFamily: Hypothetical protein YojF85 d1w07a2
not modelled
10.7
2
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: acyl-CoA oxidase C-terminal domains86 c1qrjB_
not modelled
10.7
9
PDB header: viral proteinChain: B: PDB Molecule: htlv-i capsid protein;PDBTitle: solution structure of htlv-i capsid protein
87 c2r5uD_
not modelled
10.6
17
PDB header: hydrolaseChain: D: PDB Molecule: replicative dna helicase;PDBTitle: crystal structure of the n-terminal domain of dnab helicase from2 mycobacterium tuberculosis
88 c3kvpB_
not modelled
10.6
33
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein ymzc;PDBTitle: crystal structure of uncharacterized protein ymzc precursor2 from bacillus subtilis, northeast structural genomics3 consortium target sr378a
89 d1f35a_
not modelled
10.6
21
Fold: Olfactory marker proteinSuperfamily: Olfactory marker proteinFamily: Olfactory marker protein90 d1wb8a1
not modelled
10.5
15
Fold: Long alpha-hairpinSuperfamily: Fe,Mn superoxide dismutase (SOD), N-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), N-terminal domain91 c2uvgA_
not modelled
10.4
19
PDB header: sugar-binding proteinChain: A: PDB Molecule: abc type periplasmic sugar-binding protein;PDBTitle: structure of a periplasmic oligogalacturonide binding2 protein from yersinia enterocolitica
92 d1hv8a2
not modelled
10.4
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain93 d1xr7a_
not modelled
10.3
14
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: RNA-dependent RNA-polymerase94 d1jwea_
not modelled
10.3
13
Fold: N-terminal domain of DnaB helicaseSuperfamily: N-terminal domain of DnaB helicaseFamily: N-terminal domain of DnaB helicase95 d2in1a1
not modelled
10.3
33
Fold: UBC-likeSuperfamily: UBC-likeFamily: UFC1-like96 d1ufwa_
not modelled
10.1
17
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD97 c3i1lC_
not modelled
10.1
25
PDB header: hydrolaseChain: C: PDB Molecule: hemagglutinin-esterase protein;PDBTitle: structure of porcine torovirus hemagglutinin-esterase in complex with2 its receptor
98 c1fi8E_
not modelled
9.9
36
PDB header: hydrolase/hydrolase inhibitorChain: E: PDB Molecule: ecotin;PDBTitle: rat granzyme b [n66q] complexed to ecotin [81-84 iepd]
99 d1ub4c_
not modelled
9.9
15
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: Kis/PemI addiction antidote