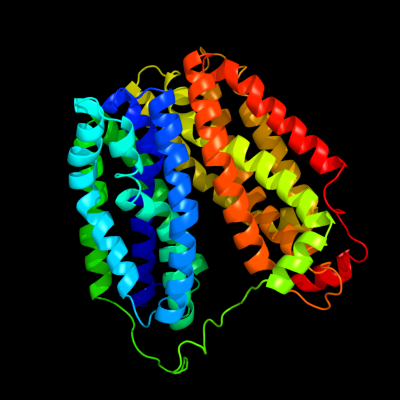
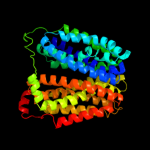
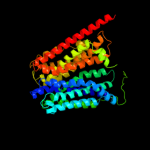
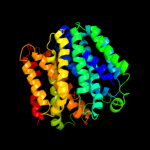
1 d1pv7a_
99.9
12
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: LacY-like proton/sugar symporter2 d1pw4a_
99.9
13
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: Glycerol-3-phosphate transporter3 c2gfpA_
99.8
12
PDB header: membrane proteinChain: A: PDB Molecule: multidrug resistance protein d;PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
4 c3o7pA_
99.8
13
PDB header: transport proteinChain: A: PDB Molecule: l-fucose-proton symporter;PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
5 c2xutC_
99.8
11
PDB header: transport proteinChain: C: PDB Molecule: proton/peptide symporter family protein;PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
6 d2f23a1
62.4
10
Fold: Long alpha-hairpinSuperfamily: GreA transcript cleavage protein, N-terminal domainFamily: GreA transcript cleavage protein, N-terminal domain7 d1grja1
50.2
24
Fold: Long alpha-hairpinSuperfamily: GreA transcript cleavage protein, N-terminal domainFamily: GreA transcript cleavage protein, N-terminal domain8 c3qnqD_
49.6
14
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
9 c1zxaB_
32.9
19
PDB header: transferaseChain: B: PDB Molecule: cgmp-dependent protein kinase 1, alpha isozyme;PDBTitle: solution structure of the coiled-coil domain of cgmp-2 dependent protein kinase ia
10 c1by0A_
29.6
28
PDB header: rna binding proteinChain: A: PDB Molecule: protein (hepatitis delta antigen);PDBTitle: n-terminal leucine-repeat region of hepatitis delta antigen
11 c2q7cC_
25.0
22
PDB header: viral proteinChain: C: PDB Molecule: fusion protein between yeast variant gcn4 andPDBTitle: crystal structure of iqn17
12 d1f6ga_
24.9
13
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels13 c3hd6A_
23.7
7
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: ammonium transporter rh type c;PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
14 d1oqwa_
20.4
13
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin15 d1xmpa_
19.7
10
Fold: Flavodoxin-likeSuperfamily: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)Family: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)16 d2h8aa1
17.4
7
Fold: MAPEG domain-likeSuperfamily: MAPEG domain-likeFamily: MAPEG domain17 d2pila_
17.1
9
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin18 c3jqhA_
13.8
24
PDB header: sugar binding proteinChain: A: PDB Molecule: c-type lectin domain family 4 member m;PDBTitle: structure of the neck region of the glycan-binding receptor2 dc-signr
19 d1ji6a3
13.3
14
Fold: Toxins' membrane translocation domainsSuperfamily: delta-Endotoxin (insectocide), N-terminal domainFamily: delta-Endotoxin (insectocide), N-terminal domain20 c2hjmB_
12.8
10
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein pf1176;PDBTitle: crystal structure of a singleton protein pf1176 from p. furiosus
21 d2ce7a1
not modelled
12.0
13
Fold: FtsH protease domain-likeSuperfamily: FtsH protease domain-likeFamily: FtsH protease domain-like22 c1a92B_
not modelled
11.8
29
PDB header: leucine zipperChain: B: PDB Molecule: delta antigen;PDBTitle: oligomerization domain of hepatitis delta antigen
23 c1m46B_
not modelled
11.7
29
PDB header: cell cycle proteinChain: B: PDB Molecule: iq4 motif from myo2p, a class v myosin;PDBTitle: crystal structure of mlc1p bound to iq4 of myo2p, a class v2 myosin
24 c2nuxB_
not modelled
11.5
35
PDB header: lyaseChain: B: PDB Molecule: 2-keto-3-deoxygluconate/2-keto-3-deoxy-6-phospho gluconatePDBTitle: 2-keto-3-deoxygluconate aldolase from sulfolobus acidocaldarius,2 native structure in p6522 at 2.5 a resolution
25 c2oqqB_
not modelled
11.5
16
PDB header: transcriptionChain: B: PDB Molecule: transcription factor hy5;PDBTitle: crystal structure of hy5 leucine zipper homodimer from2 arabidopsis thaliana
26 c3eh0C_
not modelled
10.4
8
PDB header: transferaseChain: C: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] glucosamine n-PDBTitle: crystal structure of lpxd from escherichia coli
27 d1u69a_
not modelled
9.6
27
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: 3-demethylubiquinone-9 3-methyltransferase28 c2wl2B_
not modelled
9.2
11
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase subunit a;PDBTitle: crystal structure of n-terminal domain of gyra with the2 antibiotic simocyclinone d8
29 c2xseA_
not modelled
9.2
21
PDB header: oxidoreductaseChain: A: PDB Molecule: thymine dioxygenase jbp1;PDBTitle: the structural basis for recognition of j-base containing2 dna by a novel dna-binding domain in jbp1
30 c1c94B_
not modelled
8.8
15
PDB header: gene regulationChain: B: PDB Molecule: retro-gcn4 leucine zipper;PDBTitle: reversing the sequence of the gcn4 leucine zipper does not2 affect its fold.
31 c2kncA_
not modelled
8.8
13
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
32 c1l4aE_
not modelled
8.7
22
PDB header: endocytosis/exocytosisChain: E: PDB Molecule: synaphin a;PDBTitle: x-ray structure of the neuronal complexin/snare complex2 from the squid loligo pealei
33 c3s2xB_
not modelled
8.5
23
PDB header: transferaseChain: B: PDB Molecule: acetyl-coa synthase subunit alpha;PDBTitle: structure of acetyl-coenzyme a synthase alpha subunit c-terminal2 domain
34 d2ia9a1
not modelled
8.5
7
Fold: SpoVG-likeSuperfamily: SpoVG-likeFamily: SpoVG-like35 d1ab4a_
not modelled
8.4
13
Fold: Type II DNA topoisomeraseSuperfamily: Type II DNA topoisomeraseFamily: Type II DNA topoisomerase36 c2l5gA_
not modelled
8.3
9
PDB header: transcription regulatorChain: A: PDB Molecule: g protein pathway suppressor 2;PDBTitle: co-ordinates and 1h, 13c and 15n chemical shift assignments for the2 complex of gps2 53-90 and smrt 167-207
37 c1ji6A_
not modelled
8.1
14
PDB header: toxinChain: A: PDB Molecule: pesticidial crystal protein cry3bb;PDBTitle: crystal structure of the insecticidal bacterial del2 endotoxin cry3bb1 bacillus thuringiensis
38 d2oara1
not modelled
8.1
18
Fold: Gated mechanosensitive channelSuperfamily: Gated mechanosensitive channelFamily: Gated mechanosensitive channel39 c3sokB_
not modelled
7.9
17
PDB header: cell adhesionChain: B: PDB Molecule: fimbrial protein;PDBTitle: dichelobacter nodosus pilin fima
40 d1ez4a2
not modelled
7.6
23
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain41 d1ykhb1
not modelled
7.4
8
Fold: Mediator hinge subcomplex-likeSuperfamily: Mediator hinge subcomplex-likeFamily: CSE2-like42 c2novD_
not modelled
7.4
21
PDB header: isomeraseChain: D: PDB Molecule: dna topoisomerase 4 subunit a;PDBTitle: breakage-reunion domain of s.pneumoniae topo iv: crystal2 structure of a gram-positive quinolone target
43 c2kluA_
not modelled
7.3
10
PDB header: immune system, membrane proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd4;PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
44 c1tjlD_
not modelled
7.2
0
PDB header: transcriptionChain: D: PDB Molecule: dnak suppressor protein;PDBTitle: crystal structure of transcription factor dksa from e. coli
45 c2rdcA_
not modelled
7.1
10
PDB header: lipid binding proteinChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative lipid binding protein (gsu0061) from2 geobacter sulfurreducens pca at 1.80 a resolution
46 c2kwyA_
not modelled
7.1
19
PDB header: proton transportChain: A: PDB Molecule: v-type proton atpase subunit g;PDBTitle: structure of g61-101
47 c3iz5K_
not modelled
7.1
11
PDB header: ribosomeChain: K: PDB Molecule: 60s ribosomal protein l13a (l13p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
48 c3onoA_
not modelled
7.1
14
PDB header: isomeraseChain: A: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of ribose-5-phosphate isomerase lacab_rpib from2 vibrio parahaemolyticus
49 c2kmfA_
not modelled
7.1
16
PDB header: photosynthesisChain: A: PDB Molecule: photosystem ii 11 kda protein;PDBTitle: solution structure of psb27 from cyanobacterial photosystem2 ii
50 c2y6xA_
not modelled
7.0
20
PDB header: photosynthesisChain: A: PDB Molecule: photosystem ii 11 kd protein;PDBTitle: structure of psb27 from thermosynechococcus elongatus
51 d1otsa_
not modelled
6.9
13
Fold: Clc chloride channelSuperfamily: Clc chloride channelFamily: Clc chloride channel52 c2w8aC_
not modelled
6.7
8
PDB header: membrane proteinChain: C: PDB Molecule: glycine betaine transporter betp;PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
53 d2cqea1
not modelled
6.7
10
Fold: CCCH zinc fingerSuperfamily: CCCH zinc fingerFamily: CCCH zinc finger54 c2jo8B_
not modelled
6.6
25
PDB header: transferaseChain: B: PDB Molecule: serine/threonine-protein kinase 4;PDBTitle: solution structure of c-terminal domain of human mammalian2 sterile 20-like kinase 1 (mst1)
55 d2ba0d2
not modelled
6.6
15
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like56 c2zxrA_
not modelled
6.5
12
PDB header: hydrolaseChain: A: PDB Molecule: single-stranded dna specific exonuclease recj;PDBTitle: crystal structure of recj in complex with mg2+ from thermus2 thermophilus hb8
57 d1fx0a1
not modelled
6.4
11
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase58 d2jf2a1
not modelled
6.4
13
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: UDP N-acetylglucosamine acyltransferase59 c1onvB_
not modelled
6.1
22
PDB header: transcriptionChain: B: PDB Molecule: serine phosphatase fcp1a;PDBTitle: nmr structure of a complex containing the tfiif subunit2 rap74 and the rnap ii ctd phosphatase fcp1
60 d2b5ua2
not modelled
6.1
17
Fold: Cloacin translocation domainSuperfamily: Cloacin translocation domainFamily: Cloacin translocation domain61 d1h41a2
not modelled
6.0
20
Fold: Zincin-likeSuperfamily: beta-N-acetylhexosaminidase-like domainFamily: alpha-D-glucuronidase, N-terminal domain62 d1hyha2
not modelled
5.9
10
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain63 c2q9qC_
not modelled
5.8
7
PDB header: replicationChain: C: PDB Molecule: dna replication complex gins protein psf1;PDBTitle: the crystal structure of full length human gins complex
64 c2l16A_
not modelled
5.8
15
PDB header: protein transportChain: A: PDB Molecule: sec-independent protein translocase protein tatad;PDBTitle: solution structure of bacillus subtilits tatad protein in dpc micelles
65 c2rpaA_
not modelled
5.8
24
PDB header: hydrolaseChain: A: PDB Molecule: katanin p60 atpase-containing subunit a1;PDBTitle: the solution structure of n-terminal domain of microtubule severing2 enzyme
66 c2xzrA_
not modelled
5.7
21
PDB header: cell adhesionChain: A: PDB Molecule: immunoglobulin-binding protein eibd;PDBTitle: escherichia coli immunoglobulin-binding protein eibd 391-438 fused2 to gcn4 adaptors
67 c2inrA_
not modelled
5.7
24
PDB header: isomeraseChain: A: PDB Molecule: dna topoisomerase 4 subunit a;PDBTitle: crystal structure of a 59 kda fragment of topoisomerase iv subunit a2 (grla) from staphylococcus aureus
68 c2k6iA_
not modelled
5.7
18
PDB header: structural proteinChain: A: PDB Molecule: uncharacterized protein mj0223;PDBTitle: the domain features of the peripheral stalk subunit h of the2 methanogenic a1ao atp synthase and the nmr solution3 structure of h1-47
69 c1zvuA_
not modelled
5.6
18
PDB header: isomeraseChain: A: PDB Molecule: topoisomerase iv subunit a;PDBTitle: structure of the full-length e. coli parc subunit
70 c1unyA_
not modelled
5.6
9
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
71 c2c5qE_
not modelled
5.4
23
PDB header: structural genomics,unknown functionChain: E: PDB Molecule: rraa-like protein yer010c;PDBTitle: crystal structure of yeast yer010cp
72 c3izcK_
not modelled
5.4
21
PDB header: ribosomeChain: K: PDB Molecule: 60s ribosomal protein rpl16 (l13p);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
73 c2hw2A_
not modelled
5.4
7
PDB header: transferaseChain: A: PDB Molecule: rifampin adp-ribosyl transferase;PDBTitle: crystal structure of rifampin adp-ribosyl transferase in2 complex with rifampin
74 c2rddB_
not modelled
5.3
15
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
75 d2e9xa1
not modelled
5.3
7
Fold: GINS helical bundle-likeSuperfamily: GINS helical bundle-likeFamily: PSF1 N-terminal domain-like76 c3gb8B_
not modelled
5.3
4
PDB header: transport proteinChain: B: PDB Molecule: snurportin-1;PDBTitle: crystal structure of crm1/snurportin-1 complex
77 c1nohB_
not modelled
5.3
14
PDB header: viral proteinChain: B: PDB Molecule: head morphogenesis protein;PDBTitle: the structure of bacteriophage phi29 scaffolding protein2 gp7 after prohead assembly
78 c3ei1A_
not modelled
5.3
17
PDB header: dna binding protein/dnaChain: A: PDB Molecule: dna damage-binding protein 1;PDBTitle: structure of hsddb1-drddb2 bound to a 14 bp 6-42 photoproduct containing dna-duplex
79 c1ij3B_
not modelled
5.3
8
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
80 c1ij3C_
not modelled
5.3
8
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
81 c3ilwA_
not modelled
5.3
11
PDB header: isomeraseChain: A: PDB Molecule: dna gyrase subunit a;PDBTitle: structure of dna gyrase subunit a n-terminal domain
82 d1i9za_
not modelled
5.3
22
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: Inositol polyphosphate 5-phosphatase (IPP5)83 d2fgea3
not modelled
5.3
15
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like84 c3eb7B_
not modelled
5.2
18
PDB header: toxinChain: B: PDB Molecule: insecticidal delta-endotoxin cry8ea1;PDBTitle: crystal structure of insecticidal delta-endotoxin cry8ea1 from2 bacillus thuringiensis at 2.2 angstroms resolution
85 d1gega_
not modelled
5.2
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases86 d1k1xa1
not modelled
5.2
11
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Families 57/38 glycoside transferase middle domainFamily: 4-alpha-glucanotransferase, domain 287 c3jywM_
not modelled
5.2
5
PDB header: ribosomeChain: M: PDB Molecule: 60s ribosomal protein l16(a);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
88 c1zi7C_
not modelled
5.1
10
PDB header: lipid binding proteinChain: C: PDB Molecule: kes1 protein;PDBTitle: structure of truncated yeast oxysterol binding protein osh4
89 c3ifzA_
not modelled
5.1
11
PDB header: isomeraseChain: A: PDB Molecule: dna gyrase subunit a;PDBTitle: crystal structure of the first part of the mycobacterium tuberculosis2 dna gyrase reaction core: the breakage and reunion domain at 2.7 a3 resolution
90 d1tsja_
not modelled
5.1
18
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: 3-demethylubiquinone-9 3-methyltransferase91 c2yggA_
not modelled
5.0
25
PDB header: metal binding protein/transport proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: complex of cambr and cam