| 1 | c3i4oA_ |
|
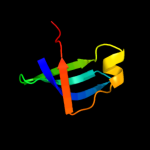 |
91.6 |
20 |
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor if-1;
PDBTitle: crystal structure of translation initiation factor 1 from2 mycobacterium tuberculosis
|
| 2 | c1zn1L_ |
|
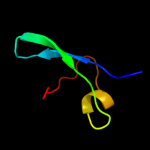 |
87.1 |
17 |
PDB header:biosynthetic/structural protein/rna
Chain: L: PDB Molecule:30s ribosomal protein s12;
PDBTitle: coordinates of rrf fitted into cryo-em map of the 70s post-2 termination complex
|
| 3 | c3fovA_ |
|
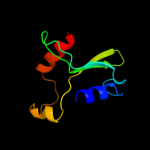 |
87.0 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0102 protein rpa0323;
PDBTitle: crystal structure of protein rpa0323 of unknown function from2 rhodopseudomonas palustris
|
| 4 | c2oqkA_ |
|
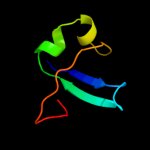 |
86.8 |
31 |
PDB header:translation
Chain: A: PDB Molecule:putative translation initiation factor eif-1a;
PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
|
| 5 | c1xdoB_ |
|
 |
85.1 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of escherichia coli polyphosphate kinase
|
| 6 | d2o8ra4 |
|
 |
84.2 |
21 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 7 | d1ah9a_ |
|
 |
83.8 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 8 | d2uubl1 |
|
 |
83.5 |
26 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 9 | d1d7qa_ |
|
 |
83.0 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 10 | d1xdpa4 |
|
 |
82.7 |
19 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 11 | c2o8rA_ |
|
 |
77.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of polyphosphate kinase from2 porphyromonas gingivalis
|
| 12 | d1jt8a_ |
|
 |
75.0 |
13 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 13 | d2qall1 |
|
 |
74.9 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 14 | c2zkql_ |
|
 |
72.0 |
21 |
PDB header:ribosomal protein/rna
Chain: L: PDB Molecule:
PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 15 | c1s1hL_ |
|
 |
70.3 |
24 |
PDB header:ribosome
Chain: L: PDB Molecule:40s ribosomal protein s23;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
|
| 16 | d1i94l_ |
|
 |
69.3 |
26 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 17 | c3m9bK_ |
|
 |
68.6 |
27 |
PDB header:chaperone
Chain: K: PDB Molecule:proteasome-associated atpase;
PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
|
| 18 | c2xzmL_ |
|
 |
68.5 |
15 |
PDB header:ribosome
Chain: L: PDB Molecule:40s ribosomal protein s12;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
|
| 19 | c2vldA_ |
|
 |
66.2 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:upf0286 protein pyrab01260;
PDBTitle: crystal structure of a repair endonuclease from pyrococcus2 abyssi
|
| 20 | c3m0zD_ |
|
 |
62.1 |
38 |
PDB header:lyase
Chain: D: PDB Molecule:putative aldolase;
PDBTitle: crystal structure of putative aldolase from klebsiella2 pneumoniae.
|
| 21 | d1hr0w_ |
|
not modelled |
59.7 |
24 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 22 | c2a5hC_ |
|
not modelled |
57.9 |
19 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
| 23 | c2dgyA_ |
|
not modelled |
53.5 |
12 |
PDB header:translation
Chain: A: PDB Molecule:mgc11102 protein;
PDBTitle: solution structure of the eukaryotic initiation factor 1a2 in mgc11102 protein
|
| 24 | d1v0wa2 |
|
not modelled |
49.1 |
17 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Phospholipase D |
| 25 | d1u0la1 |
|
not modelled |
45.8 |
16 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 26 | c3fp9E_ |
|
not modelled |
43.3 |
26 |
PDB header:hydrolase
Chain: E: PDB Molecule:proteasome-associated atpase;
PDBTitle: crystal structure of intern domain of proteasome-associated2 atpase, mycobacterium tuberculosis
|
| 27 | d2ebfx2 |
|
not modelled |
42.6 |
20 |
Fold:EreA/ChaN-like
Superfamily:EreA/ChaN-like
Family:PMT domain-like |
| 28 | c1pk8D_ |
|
not modelled |
41.6 |
19 |
PDB header:membrane protein
Chain: D: PDB Molecule:rat synapsin i;
PDBTitle: crystal structure of rat synapsin i c domain complexed to2 ca.atp
|
| 29 | d2ga5a1 |
|
not modelled |
34.5 |
23 |
Fold:N domain of copper amine oxidase-like
Superfamily:Frataxin/Nqo15-like
Family:Frataxin-like |
| 30 | d1ekga_ |
|
not modelled |
32.7 |
28 |
Fold:N domain of copper amine oxidase-like
Superfamily:Frataxin/Nqo15-like
Family:Frataxin-like |
| 31 | c3muxB_ |
|
not modelled |
31.6 |
33 |
PDB header:lyase
Chain: B: PDB Molecule:putative 4-hydroxy-2-oxoglutarate aldolase;
PDBTitle: the crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase2 from bacillus anthracis to 1.45a
|
| 32 | c3m6yA_ |
|
not modelled |
29.3 |
31 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase;
PDBTitle: structure of 4-hydroxy-2-oxoglutarate aldolase from bacillus cereus at2 1.45 a resolution.
|
| 33 | c2qs7D_ |
|
not modelled |
26.2 |
32 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative oxidoreductase of the dsre/dsrf-like2 family (sso1126) from sulfolobus solfataricus p2 at 2.09 a resolution
|
| 34 | c2pd2A_ |
|
not modelled |
25.8 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein st0148;
PDBTitle: crystal structure of (st0148) conserved hypothetical from sulfolobus2 tokodaii strain7
|
| 35 | d2p97a1 |
|
not modelled |
25.6 |
10 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Ava3068-like |
| 36 | c1nm3B_ |
|
not modelled |
24.2 |
21 |
PDB header:electron transport
Chain: B: PDB Molecule:protein hi0572;
PDBTitle: crystal structure of heamophilus influenza hybrid-prx5
|
| 37 | d1i7na2 |
|
not modelled |
23.5 |
15 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Synapsin C-terminal domain |
| 38 | d2cv4a1 |
|
not modelled |
21.1 |
22 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 39 | d1byra_ |
|
not modelled |
20.6 |
24 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Nuclease |
| 40 | c1yjxD_ |
|
not modelled |
20.1 |
19 |
PDB header:isomerase, hydrolase
Chain: D: PDB Molecule:phosphoglycerate mutase 1;
PDBTitle: crystal structure of human b type phosphoglycerate mutase
|
| 41 | c3m9pA_ |
|
not modelled |
19.6 |
13 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:male-specific lethal 3 homolog;
PDBTitle: human msl3 chromodomain bound to dna and h4k20me1 peptide
|
| 42 | c3pnxF_ |
|
not modelled |
19.0 |
33 |
PDB header:transferase
Chain: F: PDB Molecule:putative sulfurtransferase dsre;
PDBTitle: crystal structure of a putative sulfurtransferase dsre (swol_2425)2 from syntrophomonas wolfei str. goettingen at 1.92 a resolution
|
| 43 | c2r2dC_ |
|
not modelled |
18.9 |
15 |
PDB header:hydrolase
Chain: C: PDB Molecule:zn-dependent hydrolases;
PDBTitle: structure of a quorum-quenching lactonase (aiib) from agrobacterium2 tumefaciens
|
| 44 | c3s9xA_ |
|
not modelled |
18.6 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:asch domain;
PDBTitle: high resolution crystal structure of asch domain from lactobacillus2 crispatus jv v101
|
| 45 | d2rdea2 |
|
not modelled |
18.5 |
11 |
Fold:Split barrel-like
Superfamily:PilZ domain-like
Family:PilZ domain-associated domain |
| 46 | d1e32a1 |
|
not modelled |
17.7 |
19 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Cdc48 N-terminal domain-like |
| 47 | d2okfa1 |
|
not modelled |
17.2 |
20 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:XisH-like |
| 48 | d1jx7a_ |
|
not modelled |
17.2 |
13 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrEF-like |
| 49 | d1xdpa3 |
|
not modelled |
17.1 |
14 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 50 | c3dnxA_ |
|
not modelled |
16.9 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein spo1766;
PDBTitle: spo1766 protein of unknown function from silicibacter pomeroyi.
|
| 51 | d2inba1 |
|
not modelled |
16.4 |
20 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:XisH-like |
| 52 | d1o12a1 |
|
not modelled |
15.2 |
21 |
Fold:Composite domain of metallo-dependent hydrolases
Superfamily:Composite domain of metallo-dependent hydrolases
Family:N-acetylglucosamine-6-phosphate deacetylase, NagA |
| 53 | c3h43F_ |
|
not modelled |
14.9 |
17 |
PDB header:hydrolase
Chain: F: PDB Molecule:proteasome-activating nucleotidase;
PDBTitle: n-terminal domain of the proteasome-activating nucleotidase2 of methanocaldococcus jannaschii
|
| 54 | c3m9qA_ |
|
not modelled |
14.8 |
9 |
PDB header:dna binding protein
Chain: A: PDB Molecule:protein male-specific lethal-3;
PDBTitle: drosophila msl3 chromodomain
|
| 55 | c2br6A_ |
|
not modelled |
14.7 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:aiia-like protein;
PDBTitle: crystal structure of quorum-quenching n-acyl homoserine2 lactone lactonase
|
| 56 | d1g94a2 |
|
not modelled |
14.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 57 | c3dcyA_ |
|
not modelled |
13.7 |
19 |
PDB header:apoptosis regulator
Chain: A: PDB Molecule:regulator protein;
PDBTitle: crystal structure a tp53-induced glycolysis and apoptosis2 regulator protein from homo sapiens.
|
| 58 | d2fqla1 |
|
not modelled |
12.8 |
19 |
Fold:N domain of copper amine oxidase-like
Superfamily:Frataxin/Nqo15-like
Family:Frataxin-like |
| 59 | c3o27B_ |
|
not modelled |
12.6 |
19 |
PDB header:dna binding protein
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: the crystal structure of c68 from the hybrid virus-plasmid pssvx
|
| 60 | d1l1sa_ |
|
not modelled |
11.5 |
20 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrEF-like |
| 61 | c3tinA_ |
|
not modelled |
11.5 |
33 |
PDB header:ligase
Chain: A: PDB Molecule:ttl protein;
PDBTitle: tubulin tyrosine ligase
|
| 62 | d1pk8a2 |
|
not modelled |
11.4 |
23 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Synapsin C-terminal domain |
| 63 | c2wg6L_ |
|
not modelled |
11.4 |
16 |
PDB header:transcription,hydrolase
Chain: L: PDB Molecule:general control protein gcn4,
PDBTitle: proteasome-activating nucleotidase (pan) n-domain (57-134)2 from archaeoglobus fulgidus fused to gcn4, p61a mutant
|
| 64 | c2fb6A_ |
|
not modelled |
11.0 |
30 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: structure of conserved protein of unknown function bt1422 from2 bacteroides thetaiotaomicron
|
| 65 | c2wagA_ |
|
not modelled |
10.7 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:lysozyme, putative;
PDBTitle: the structure of a family 25 glycosyl hydrolase from2 bacillus anthracis.
|
| 66 | c2qpqC_ |
|
not modelled |
10.5 |
13 |
PDB header:transport protein
Chain: C: PDB Molecule:protein bug27;
PDBTitle: structure of bug27 from bordetella pertussis
|
| 67 | d1qnia1 |
|
not modelled |
10.3 |
15 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Nitrosocyanin |
| 68 | c3e9eB_ |
|
not modelled |
10.0 |
9 |
PDB header:hydrolase
Chain: B: PDB Molecule:zgc:56074;
PDBTitle: structure of full-length h11a mutant form of tigar from danio rerio
|
| 69 | c3aj3A_ |
|
not modelled |
10.0 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:4-pyridoxolactonase;
PDBTitle: crystal structure of selenomethionine substituted 4-pyridoxolactonase2 from mesorhizobium loti
|
| 70 | c3i38H_ |
|
not modelled |
9.9 |
17 |
PDB header:chaperone
Chain: H: PDB Molecule:putative chaperone dnaj;
PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
|
| 71 | c2rcnA_ |
|
not modelled |
9.6 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable gtpase engc;
PDBTitle: crystal structure of the ribosomal interacting gtpase yjeq from the2 enterobacterial species salmonella typhimurium.
|
| 72 | d2gjpa2 |
|
not modelled |
9.3 |
6 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 73 | d1ea9c3 |
|
not modelled |
8.9 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 74 | d1s98a_ |
|
not modelled |
8.9 |
11 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 75 | d1t62a_ |
|
not modelled |
8.5 |
28 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:Hypothetical protein EF3133 |
| 76 | c2vpmB_ |
|
not modelled |
8.5 |
21 |
PDB header:ligase
Chain: B: PDB Molecule:trypanothione synthetase;
PDBTitle: trypanothione synthetase
|
| 77 | c1u0lB_ |
|
not modelled |
8.4 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable gtpase engc;
PDBTitle: crystal structure of yjeq from thermotoga maritima
|
| 78 | d1nm3a2 |
|
not modelled |
8.4 |
17 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 79 | d1pkla1 |
|
not modelled |
7.8 |
21 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 80 | c1wn4A_ |
|
not modelled |
7.7 |
36 |
PDB header:plant protein
Chain: A: PDB Molecule:vontr protein;
PDBTitle: nmr structure of vontr
|
| 81 | d1ua7a2 |
|
not modelled |
7.5 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 82 | d1a3xa1 |
|
not modelled |
7.4 |
27 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 83 | d3e9va1 |
|
not modelled |
7.3 |
18 |
Fold:BTG domain-like
Superfamily:BTG domain-like
Family:BTG domain-like |
| 84 | c3dhuC_ |
|
not modelled |
7.0 |
12 |
PDB header:hydrolase
Chain: C: PDB Molecule:alpha-amylase;
PDBTitle: crystal structure of an alpha-amylase from lactobacillus2 plantarum
|
| 85 | c2yv5A_ |
|
not modelled |
7.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:yjeq protein;
PDBTitle: crystal structure of yjeq from aquifex aeolicus
|
| 86 | c3bmwA_ |
|
not modelled |
6.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:cyclomaltodextrin glucanotransferase;
PDBTitle: cyclodextrin glycosyl transferase from thermoanerobacterium2 thermosulfurigenes em1 mutant s77p complexed with a maltoheptaose3 inhibitor
|
| 87 | c1bplA_ |
|
not modelled |
6.6 |
9 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:alpha-1,4-glucan-4-glucanohydrolase;
PDBTitle: glycosyltransferase
|
| 88 | d1lkxa_ |
|
not modelled |
6.4 |
8 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Motor proteins |
| 89 | d1zbsa2 |
|
not modelled |
6.4 |
15 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:BadF/BadG/BcrA/BcrD-like |
| 90 | d1cw0a_ |
|
not modelled |
6.3 |
16 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Very short patch repair (VSR) endonuclease |
| 91 | c2zfnA_ |
|
not modelled |
6.2 |
40 |
PDB header:transferase
Chain: A: PDB Molecule:regulator of ty1 transposition protein 109;
PDBTitle: self-acetylation mediated histone h3 lysine 56 acetylation by rtt109
|
| 92 | c3d35A_ |
|
not modelled |
6.2 |
40 |
PDB header:transferase
Chain: A: PDB Molecule:regulator of ty1 transposition protein 109;
PDBTitle: crystal structure of rtt109-ac-coa complex
|
| 93 | d2z15a1 |
|
not modelled |
5.9 |
14 |
Fold:BTG domain-like
Superfamily:BTG domain-like
Family:BTG domain-like |
| 94 | c3i38L_ |
|
not modelled |
5.9 |
18 |
PDB header:chaperone
Chain: L: PDB Molecule:putative chaperone dnaj;
PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
|
| 95 | c3cz7A_ |
|
not modelled |
5.9 |
40 |
PDB header:replication
Chain: A: PDB Molecule:regulator of ty1 transposition protein 109;
PDBTitle: molecular basis for the autoregulation of the protein acetyl2 transferase rtt109
|
| 96 | c1i7nA_ |
|
not modelled |
5.8 |
15 |
PDB header:neuropeptide
Chain: A: PDB Molecule:synapsin ii;
PDBTitle: crystal structure analysis of the c domain of synapsin ii2 from rat brain
|
| 97 | c1ud8A_ |
|
not modelled |
5.8 |
0 |
PDB header:hydrolase
Chain: A: PDB Molecule:amylase;
PDBTitle: crystal structure of amyk38 with lithium ion
|
| 98 | c1sg3A_ |
|
not modelled |
5.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:allantoicase;
PDBTitle: structure of allantoicase
|
| 99 | c2nw0B_ |
|
not modelled |
5.7 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:plyb;
PDBTitle: crystal structure of a lysin
|

