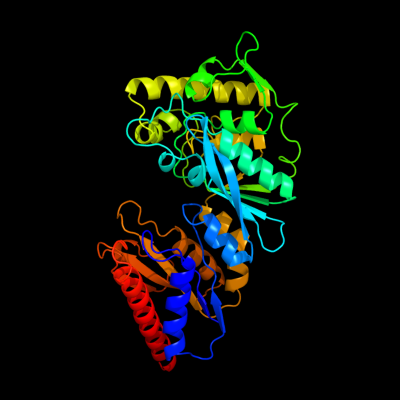
| 1 | c3cwcB_
|
|
 |
100.0 |
56 |
PDB header:transferase
Chain: B: PDB Molecule:putative glycerate kinase 2;
PDBTitle: crystal structure of putative glycerate kinase 2 from salmonella2 typhimurium lt2
|
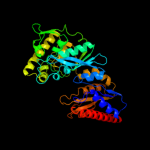
| 2 | d1to6a_
|
|
 |
100.0 |
39 |
Fold:Glycerate kinase I
Superfamily:Glycerate kinase I
Family:Glycerate kinase I |
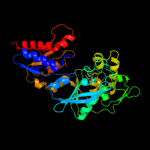
| 3 | d1e0ta2
|
|
 |
95.1 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 4 | d1liua2
|
|
 |
94.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 5 | d1pkla2
|
|
 |
93.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 6 | d1a3xa2
|
|
 |
89.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 7 | d1vb5a_
|
|
 |
88.7 |
16 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 8 | d1pswa_
|
|
 |
87.3 |
15 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:ADP-heptose LPS heptosyltransferase II |
| 9 | d2g50a2
|
|
 |
87.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 10 | c1w2wJ_
|
|
 |
84.6 |
22 |
PDB header:isomerase
Chain: J: PDB Molecule:5-methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of yeast ypr118w, a methylthioribose-1-2 phosphate isomerase related to regulatory eif2b subunits
|
| 11 | c3ecsD_
|
|
 |
84.2 |
24 |
PDB header:translation
Chain: D: PDB Molecule:translation initiation factor eif-2b subunit
PDBTitle: crystal structure of human eif2b alpha
|
| 12 | d1ydga_
|
|
 |
83.9 |
15 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 13 | c1w5fA_
|
|
 |
83.5 |
20 |
PDB header:cell division
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: ftsz, t7 mutated, domain swapped (t. maritima)
|
| 14 | d1t5oa_
|
|
 |
82.7 |
29 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 15 | d2vapa1
|
|
 |
82.6 |
22 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 16 | d1rq2a1
|
|
 |
82.1 |
24 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 17 | c3a11D_
|
|
 |
81.6 |
18 |
PDB header:isomerase
Chain: D: PDB Molecule:translation initiation factor eif-2b, delta
PDBTitle: crystal structure of ribose-1,5-bisphosphate isomerase from2 thermococcus kodakaraensis kod1
|
| 18 | c1a3wB_
|
|
 |
80.5 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase;
PDBTitle: pyruvate kinase from saccharomyces cerevisiae complexed with fbp, pg,2 mn2+ and k+
|
| 19 | c1w59B_
|
|
 |
78.7 |
21 |
PDB header:cell division
Chain: B: PDB Molecule:cell division protein ftsz homolog 1;
PDBTitle: ftsz dimer, empty (m. jannaschii)
|
| 20 | c2yvkA_
|
|
 |
75.8 |
30 |
PDB header:isomerase
Chain: A: PDB Molecule:methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
|
| 21 | d1t9ka_ |
|
not modelled |
75.7 |
30 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 22 | c2h1fB_ |
|
not modelled |
74.2 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:lipopolysaccharide heptosyltransferase-1;
PDBTitle: e. coli heptosyltransferase waac with adp
|
| 23 | d2a0ua1 |
|
not modelled |
73.9 |
21 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 24 | c1aqfB_ |
|
not modelled |
73.4 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase;
PDBTitle: pyruvate kinase from rabbit muscle with mg, k, and l-2 phospholactate
|
| 25 | c1t5aB_ |
|
not modelled |
71.7 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase, m2 isozyme;
PDBTitle: human pyruvate kinase m2
|
| 26 | c2vgbB_ |
|
not modelled |
71.6 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase isozymes r/l;
PDBTitle: human erythrocyte pyruvate kinase
|
| 27 | d1xi3a_ |
|
not modelled |
71.1 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
| 28 | d1izca_ |
|
not modelled |
69.2 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:HpcH/HpaI aldolase |
| 29 | c1izcA_ |
|
not modelled |
69.2 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:macrophomate synthase intermolecular diels-alderase;
PDBTitle: crystal structure analysis of macrophomate synthase
|
| 30 | c3eoeC_ |
|
not modelled |
69.1 |
23 |
PDB header:transferase
Chain: C: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pyruvate kinase from toxoplasma gondii, 55.m00007
|
| 31 | c2q1yB_ |
|
not modelled |
68.5 |
22 |
PDB header:cell cycle, signaling protein
Chain: B: PDB Molecule:cell division protein ftsz;
PDBTitle: crystal structure of cell division protein ftsz from mycobacterium2 tuberculosis in complex with gtp-gamma-s
|
| 32 | c3e0vB_ |
|
not modelled |
68.4 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pyruvate kinase from leishmania mexicana in2 complex with sulphate ions
|
| 33 | c2rhoB_ |
|
not modelled |
67.1 |
22 |
PDB header:cell cycle
Chain: B: PDB Molecule:cell division protein ftsz;
PDBTitle: synthetic gene encoded bacillus subtilis ftsz ncs dimer with2 bound gdp and gtp-gamma-s
|
| 34 | c1pklB_ |
|
not modelled |
65.6 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:protein (pyruvate kinase);
PDBTitle: the structure of leishmania pyruvate kinase
|
| 35 | c3t07D_ |
|
not modelled |
65.5 |
18 |
PDB header:transferase/transferase inhibitor
Chain: D: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of s. aureus pyruvate kinase in complex with a2 naturally occurring bis-indole alkaloid
|
| 36 | d1wv2a_ |
|
not modelled |
65.4 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
| 37 | d1ofua1 |
|
not modelled |
65.2 |
24 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 38 | c1e0tD_ |
|
not modelled |
64.6 |
13 |
PDB header:phosphotransferase
Chain: D: PDB Molecule:pyruvate kinase;
PDBTitle: r292d mutant of e. coli pyruvate kinase
|
| 39 | c2ppvA_ |
|
not modelled |
64.4 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a protein belonging to the upf0052 (se_0549) from2 staphylococcus epidermidis atcc 12228 at 2.00 a resolution
|
| 40 | c3tovB_ |
|
not modelled |
63.4 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyl transferase family 9;
PDBTitle: the crystal structure of the glycosyl transferase family 9 from2 veillonella parvula dsm 2008
|
| 41 | c2p0yA_ |
|
not modelled |
63.0 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein lp_0780;
PDBTitle: crystal structure of q88yi3_lacpl from lactobacillus2 plantarum. northeast structural genomics consortium target3 lpr6
|
| 42 | c2f9iD_ |
|
not modelled |
62.7 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl
PDBTitle: crystal structure of the carboxyltransferase subunit of acc2 from staphylococcus aureus
|
| 43 | c1ofuB_ |
|
not modelled |
62.7 |
24 |
PDB header:bacterial cell division inhibitor
Chain: B: PDB Molecule:cell division protein ftsz;
PDBTitle: crystal structure of sula:ftsz from pseudomonas aeruginosa
|
| 44 | d2f9yb1 |
|
not modelled |
61.3 |
15 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 45 | c2f9yB_ |
|
not modelled |
61.3 |
15 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl transferase subunit
PDBTitle: the crystal structure of the carboxyltransferase subunit of acc from2 escherichia coli
|
| 46 | d1ecfa1 |
|
not modelled |
59.7 |
12 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 47 | c2hjgA_ |
|
not modelled |
59.1 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:gtp-binding protein enga;
PDBTitle: the crystal structure of the b. subtilis yphc gtpase in2 complex with gdp
|
| 48 | c2vxyA_ |
|
not modelled |
58.4 |
25 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: the structure of ftsz from bacillus subtilis at 1.7a2 resolution
|
| 49 | c3hbmA_ |
|
not modelled |
56.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:udp-sugar hydrolase;
PDBTitle: crystal structure of pseg from campylobacter jejuni
|
| 50 | c2e28A_ |
|
not modelled |
56.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure analysis of pyruvate kinase from bacillus2 stearothermophilus
|
| 51 | d2hzba1 |
|
not modelled |
55.4 |
18 |
Fold:CofD-like
Superfamily:CofD-like
Family:CofD-like |
| 52 | d1vima_ |
|
not modelled |
55.3 |
13 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
| 53 | c3ma8A_ |
|
not modelled |
55.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of cgd1_2040, a pyruvate kinase from cryptosporidium2 parvum
|
| 54 | d2naca2 |
|
not modelled |
53.1 |
12 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 55 | c2ze3A_ |
|
not modelled |
53.0 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
| 56 | c2v82A_ |
|
not modelled |
52.9 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase;
PDBTitle: kdpgal complexed to kdpgal
|
| 57 | d1pjqa1 |
|
not modelled |
52.1 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
| 58 | c2vawA_ |
|
not modelled |
51.4 |
23 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: ftsz pseudomonas aeruginosa gdp
|
| 59 | c1pixB_ |
|
not modelled |
51.1 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:glutaconyl-coa decarboxylase a subunit;
PDBTitle: crystal structure of the carboxyltransferase subunit of the2 bacterial ion pump glutaconyl-coenzyme a decarboxylase
|
| 60 | d1f8ya_ |
|
not modelled |
50.8 |
5 |
Fold:Flavodoxin-like
Superfamily:N-(deoxy)ribosyltransferase-like
Family:N-deoxyribosyltransferase |
| 61 | d1q7ra_ |
|
not modelled |
50.0 |
15 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 62 | c2bf9A_ |
|
not modelled |
49.9 |
26 |
PDB header:hormone
Chain: A: PDB Molecule:pancreatic hormone;
PDBTitle: anisotropic refinement of avian (turkey) pancreatic2 polypeptide at 0.99 angstroms resolution.
|
| 63 | c3trjC_ |
|
not modelled |
49.9 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphoheptose isomerase;
PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
|
| 64 | d1diha1 |
|
not modelled |
49.6 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 65 | d1vrga1 |
|
not modelled |
49.6 |
31 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 66 | d1w5fa1 |
|
not modelled |
49.6 |
19 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
| 67 | d1j0aa_ |
|
not modelled |
49.2 |
14 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 68 | c4a1dG_ |
|
not modelled |
49.0 |
15 |
PDB header:ribosome
Chain: G: PDB Molecule:rpl30;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
|
| 69 | c3na3A_ |
|
not modelled |
48.8 |
25 |
PDB header:protein binding
Chain: A: PDB Molecule:dna mismatch repair protein mlh1;
PDBTitle: mutl protein homolog 1 isoform 1 from homo sapiens
|
| 70 | c3ct7E_ |
|
not modelled |
48.1 |
12 |
PDB header:isomerase
Chain: E: PDB Molecule:d-allulose-6-phosphate 3-epimerase;
PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
|
| 71 | d1pixa2 |
|
not modelled |
47.5 |
20 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 72 | d1t0kb_ |
|
not modelled |
47.2 |
13 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 73 | c1ronA_ |
|
not modelled |
47.2 |
22 |
PDB header:neuropeptide
Chain: A: PDB Molecule:neuropeptide y;
PDBTitle: nmr solution structure of human neuropeptide y
|
| 74 | c1tz5A_ |
|
not modelled |
46.8 |
30 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:chimera of pancreatic hormone and neuropeptide y;
PDBTitle: [pnpy19-23]-hpp bound to dpc micelles
|
| 75 | d1p3da1 |
|
not modelled |
45.3 |
15 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 76 | d1w41a1 |
|
not modelled |
44.8 |
21 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 77 | c3pqeD_ |
|
not modelled |
44.4 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: crystal structure of l-lactate dehydrogenase from bacillus subtilis2 with h171c mutation
|
| 78 | d1on3a1 |
|
not modelled |
44.0 |
24 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 79 | c3khdC_ |
|
not modelled |
43.9 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pff1300w.
|
| 80 | c1drwA_ |
|
not modelled |
43.8 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: escherichia coli dhpr/nhdh complex
|
| 81 | d1j6ua1 |
|
not modelled |
43.7 |
9 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 82 | d1dxea_ |
|
not modelled |
42.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:HpcH/HpaI aldolase |
| 83 | c1xnwD_ |
|
not modelled |
42.3 |
27 |
PDB header:ligase
Chain: D: PDB Molecule:propionyl-coa carboxylase complex b subunit;
PDBTitle: acyl-coa carboxylase beta subunit from s. coelicolor (pccb),2 apo form #2, mutant d422i
|
| 84 | c3o63B_ |
|
not modelled |
42.3 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:probable thiamine-phosphate pyrophosphorylase;
PDBTitle: crystal structure of thiamin phosphate synthase from mycobacterium2 tuberculosis
|
| 85 | c1x0uB_ |
|
not modelled |
42.2 |
29 |
PDB header:lyase
Chain: B: PDB Molecule:hypothetical methylmalonyl-coa decarboxylase alpha subunit;
PDBTitle: crystal structure of the carboxyl transferase subunit of putative pcc2 of sulfolobus tokodaii
|
| 86 | c3glmD_ |
|
not modelled |
41.4 |
23 |
PDB header:lyase
Chain: D: PDB Molecule:glutaconyl-coa decarboxylase subunit a;
PDBTitle: glutaconyl-coa decarboxylase a subunit from clostridium2 symbiosum co-crystallized with crotonyl-coa
|
| 87 | c2dezA_ |
|
not modelled |
41.3 |
26 |
PDB header:neuropeptide
Chain: A: PDB Molecule:peptide yy;
PDBTitle: structure of human pyy
|
| 88 | c3bzrA_ |
|
not modelled |
41.3 |
15 |
PDB header:membrane protein, protein transport
Chain: A: PDB Molecule:escu;
PDBTitle: crystal structure of escu c-terminal domain with n262d mutation, space2 group p 41 21 2
|
| 89 | d3bzra1 |
|
not modelled |
41.3 |
15 |
Fold:EscU C-terminal domain-like
Superfamily:EscU C-terminal domain-like
Family:EscU C-terminal domain-like |
| 90 | d1ycga1 |
|
not modelled |
40.3 |
32 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 91 | d1k9vf_ |
|
not modelled |
40.2 |
13 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 92 | c3qz6A_ |
|
not modelled |
40.0 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:hpch/hpai aldolase;
PDBTitle: the crystal structure of hpch/hpai aldolase from desulfitobacterium2 hafniense dcb-2
|
| 93 | d2csua2 |
|
not modelled |
40.0 |
24 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 94 | c3n6rF_ |
|
not modelled |
39.9 |
29 |
PDB header:ligase
Chain: F: PDB Molecule:propionyl-coa carboxylase, beta subunit;
PDBTitle: crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
|
| 95 | c1zosE_ |
|
not modelled |
39.3 |
19 |
PDB header:hydrolase
Chain: E: PDB Molecule:5'-methylthioadenosine / s-adenosylhomocysteine
PDBTitle: structure of 5'-methylthionadenosine/s-adenosylhomocysteine2 nucleosidase from s. pneumoniae with a transition-state3 inhibitor mt-imma
|
| 96 | c2yybA_ |
|
not modelled |
38.7 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ttha1606;
PDBTitle: crystal structure of ttha1606 from thermus thermophilus hb8
|
| 97 | c2r6r1_ |
|
not modelled |
38.5 |
20 |
PDB header:cell cycle
Chain: 1: PDB Molecule:cell division protein ftsz;
PDBTitle: aquifex aeolicus ftsz
|
| 98 | d2a7sa1 |
|
not modelled |
38.2 |
31 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 99 | c1on3E_ |
|
not modelled |
38.0 |
24 |
PDB header:transferase
Chain: E: PDB Molecule:methylmalonyl-coa carboxyltransferase 12s
PDBTitle: transcarboxylase 12s crystal structure: hexamer assembly2 and substrate binding to a multienzyme core (with3 methylmalonyl-coenzyme a and methylmalonic acid bound)
|
| 100 | d1xnya1 |
|
not modelled |
37.7 |
27 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 101 | d1nmpa_ |
|
not modelled |
37.6 |
24 |
Fold:NIF3 (NGG1p interacting factor 3)-like
Superfamily:NIF3 (NGG1p interacting factor 3)-like
Family:NIF3 (NGG1p interacting factor 3)-like |
| 102 | c1vrgE_ |
|
not modelled |
37.6 |
35 |
PDB header:ligase
Chain: E: PDB Molecule:propionyl-coa carboxylase, beta subunit;
PDBTitle: crystal structure of propionyl-coa carboxylase, beta subunit (tm0716)2 from thermotoga maritima at 2.30 a resolution
|
| 103 | c3t7yB_ |
|
not modelled |
37.5 |
18 |
PDB header:protein transport
Chain: B: PDB Molecule:yop proteins translocation protein u;
PDBTitle: structure of an autocleavage-inactive mutant of the cytoplasmic domain2 of ct091, the yscu homologue of chlamydia trachomatis
|
| 104 | d2nv0a1 |
|
not modelled |
37.5 |
9 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 105 | d1jvna2 |
|
not modelled |
37.2 |
17 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 106 | c2yxbA_ |
|
not modelled |
36.9 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
| 107 | c2zkiH_ |
|
not modelled |
36.9 |
16 |
PDB header:transcription
Chain: H: PDB Molecule:199aa long hypothetical trp repressor binding
PDBTitle: crystal structure of hypothetical trp repressor binding2 protein from sul folobus tokodaii (st0872)
|
| 108 | c2q7xA_ |
|
not modelled |
36.8 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:upf0052 protein sp_1565;
PDBTitle: crystal structure of a putative phospho transferase (sp_1565) from2 streptococcus pneumoniae tigr4 at 2.00 a resolution
|
| 109 | c3hgmD_ |
|
not modelled |
36.7 |
18 |
PDB header:signaling protein
Chain: D: PDB Molecule:universal stress protein tead;
PDBTitle: universal stress protein tead from the trap transporter2 teaabc of halomonas elongata
|
| 110 | c3ceuA_ |
|
not modelled |
36.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:thiamine phosphate pyrophosphorylase;
PDBTitle: crystal structure of thiamine phosphate pyrophosphorylase2 (bt_0647) from bacteroides thetaiotaomicron. northeast3 structural genomics consortium target btr268
|
| 111 | c3oirA_ |
|
not modelled |
36.1 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:sulfate transporter sulfate transporter family protein;
PDBTitle: crystal structure of sulfate transporter family protein from wolinella2 succinogenes
|
| 112 | d2ffea1 |
|
not modelled |
35.4 |
12 |
Fold:CofD-like
Superfamily:CofD-like
Family:CofD-like |
| 113 | c2zklA_ |
|
not modelled |
35.3 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:capsular polysaccharide synthesis enzyme cap5f;
PDBTitle: crystal structure of capsular polysaccharide assembling protein capf2 from staphylococcus aureus
|
| 114 | c2bg5C_ |
|
not modelled |
35.1 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:phosphoenolpyruvate-protein kinase;
PDBTitle: crystal structure of the phosphoenolpyruvate-binding enzyme2 i-domain from the thermoanaerobacter tengcongensis pep:3 sugar phosphotransferase system (pts)
|
| 115 | c3h4lB_ |
|
not modelled |
35.0 |
22 |
PDB header:dna binding protein, protein binding
Chain: B: PDB Molecule:dna mismatch repair protein pms1;
PDBTitle: crystal structure of n terminal domain of a dna repair protein
|
| 116 | d2bo1a1 |
|
not modelled |
34.7 |
17 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 117 | d1vhca_ |
|
not modelled |
34.2 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 118 | c2hwgA_ |
|
not modelled |
33.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of phosphorylated enzyme i of the2 phosphoenolpyruvate:sugar phosphotransferase system
|
| 119 | c2v5jB_ |
|
not modelled |
33.4 |
7 |
PDB header:lyase
Chain: B: PDB Molecule:2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase;
PDBTitle: apo class ii aldolase hpch
|
| 120 | c3f6sI_ |
|
not modelled |
33.4 |
21 |
PDB header:electron transport
Chain: I: PDB Molecule:flavodoxin;
PDBTitle: desulfovibrio desulfuricans (atcc 29577) oxidized flavodoxin2 alternate conformers
|