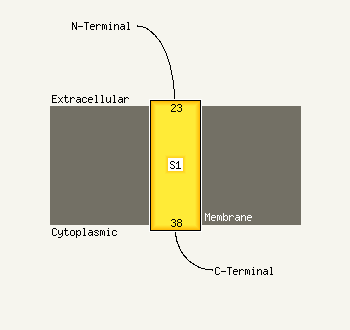
1 c2rddB_
99.1
100
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
2 c2j5uB_
88.7
27
PDB header: cell shape regulationChain: B: PDB Molecule: mrec protein;PDBTitle: mrec lysteria monocytogenes
3 c2qf4A_
88.1
35
PDB header: structural proteinChain: A: PDB Molecule: cell shape determining protein mrec;PDBTitle: high resolution structure of the major periplasmic domain from the2 cell shape-determining filament mrec (orthorhombic form)
4 d2do3a1
83.5
26
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: SPT5 KOW domain-like5 d1pkma1
77.4
12
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain6 d2g50a1
76.9
12
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain7 d1a3xa1
73.8
14
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain8 d1pkla1
72.7
9
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain9 d1liua1
72.6
12
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain10 c1kq1W_
70.4
6
PDB header: translationChain: W: PDB Molecule: host factor for q beta;PDBTitle: 1.55 a crystal structure of the pleiotropic translational2 regulator, hfq
11 c3qhsD_
68.5
18
PDB header: rna binding proteinChain: D: PDB Molecule: protein hfq;PDBTitle: crystal structure of full-length hfq from escherichia coli
12 d1hk9a_
67.5
18
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Pleiotropic translational regulator Hfq13 c2vv5D_
64.6
26
PDB header: membrane proteinChain: D: PDB Molecule: small-conductance mechanosensitive channel;PDBTitle: the open structure of mscs
14 d1u1sa1
63.3
18
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Pleiotropic translational regulator Hfq15 d2oara1
60.6
16
Fold: Gated mechanosensitive channelSuperfamily: Gated mechanosensitive channelFamily: Gated mechanosensitive channel16 c2e70A_
56.2
15
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the fifth kow motif of human2 transcription elongation factor spt5
17 d2vv5a1
56.2
26
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Mechanosensitive channel protein MscS (YggB), middle domain18 d2cp6a1
55.4
26
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain19 c2qqsB_
54.5
24
PDB header: oxidoreductaseChain: B: PDB Molecule: jmjc domain-containing histone demethylationPDBTitle: jmjd2a tandem tudor domains in complex with a trimethylated2 histone h4-k20 peptide
20 c2e6zA_
52.5
26
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
21 c3hsbB_
not modelled
51.1
18
PDB header: rna binding protein/rnaChain: B: PDB Molecule: protein hfq;PDBTitle: crystal structure of ymah (hfq) from bacillus subtilis in complex with2 an rna aptamer
22 c2l8kA_
not modelled
51.0
17
PDB header: viral proteinChain: A: PDB Molecule: non-structural protein 7;PDBTitle: nmr structure of the arterivirus nonstructural protein 7 alpha (nsp72 alpha)
23 c2equA_
not modelled
49.8
18
PDB header: protein bindingChain: A: PDB Molecule: phd finger protein 20-like 1;PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1
24 c3qiiA_
not modelled
49.5
20
PDB header: transcription regulatorChain: A: PDB Molecule: phd finger protein 20;PDBTitle: crystal structure of tudor domain 2 of human phd finger protein 20
25 c3m9bK_
not modelled
49.4
7
PDB header: chaperoneChain: K: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
26 d3bzka4
not modelled
47.7
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like27 c2oarA_
not modelled
47.5
16
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: mechanosensitive channel of large conductance (mscl)
28 d1e0ta1
not modelled
43.7
17
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain29 c1rl2A_
not modelled
43.5
20
PDB header: ribosomal proteinChain: A: PDB Molecule: protein (ribosomal protein l2);PDBTitle: ribosomal protein l2 rna-binding domain from bacillus2 stearothermophilus
30 c3nx6A_
not modelled
41.4
25
PDB header: chaperoneChain: A: PDB Molecule: 10kda chaperonin;PDBTitle: crystal structure of co-chaperonin, groes (xoo4289) from xanthomonas2 oryzae pv. oryzae kacc10331
31 c2xdpA_
not modelled
38.9
15
PDB header: oxidoreductaseChain: A: PDB Molecule: lysine-specific demethylase 4c;PDBTitle: crystal structure of the tudor domain of human jmjd2c
32 d2j01d1
not modelled
38.0
13
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: C-terminal domain of ribosomal protein L233 c4a1cA_
not modelled
36.6
17
PDB header: ribosomeChain: A: PDB Molecule: rpl8;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
34 c2khiA_
not modelled
36.3
16
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 4 of the e. coli ribosomal2 protein s1
35 d1pw4a_
not modelled
36.1
10
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: Glycerol-3-phosphate transporter36 c2jwaA_
not modelled
35.5
46
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
37 c3d5bD_
not modelled
34.7
13
PDB header: ribosomeChain: D: PDB Molecule: 50s ribosomal protein l2;PDBTitle: structural basis for translation termination on the 70s ribosome. this2 file contains the 50s subunit of one 70s ribosome. the entire crystal3 structure contains two 70s ribosomes as described in remark 400.
38 c2ivwA_
not modelled
33.9
15
PDB header: lipoproteinChain: A: PDB Molecule: pilp pilot protein;PDBTitle: the solution structure of a domain from the neisseria2 meningitidis pilp pilot protein.
39 c2dzcA_
not modelled
33.7
13
PDB header: ligaseChain: A: PDB Molecule: biotin--[acetyl-coa-carboxylase] ligase;PDBTitle: crystal structure of biotin protein ligase from pyrococcus2 horikoshii, mutation r48a
40 d2qamc1
not modelled
32.4
13
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: C-terminal domain of ribosomal protein L241 d1aono_
not modelled
31.1
21
Fold: GroES-likeSuperfamily: GroES-likeFamily: GroES42 d1p3ha_
not modelled
30.3
25
Fold: GroES-likeSuperfamily: GroES-likeFamily: GroES43 d2isba1
not modelled
30.2
25
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: FumA C-terminal domain-likeFamily: FumA C-terminal domain-like44 d2ar1a1
not modelled
28.7
27
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like45 c1aqfB_
not modelled
28.0
15
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase;PDBTitle: pyruvate kinase from rabbit muscle with mg, k, and l-2 phospholactate
46 c1kqsA_
not modelled
27.8
11
PDB header: ribosomeChain: A: PDB Molecule: ribosomal protein l2;PDBTitle: the haloarcula marismortui 50s complexed with a2 pretranslocational intermediate in protein synthesis
47 d1vqot1
not modelled
27.3
31
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e48 d1we3o_
not modelled
27.0
19
Fold: GroES-likeSuperfamily: GroES-likeFamily: GroES49 d2zjrr1
not modelled
26.7
23
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e50 d1rl2a1
not modelled
26.6
21
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: C-terminal domain of ribosomal protein L251 c2pqaB_
not modelled
26.5
19
PDB header: replicationChain: B: PDB Molecule: replication protein a 14 kda subunit;PDBTitle: crystal structure of full-length human rpa 14/32 heterodimer
52 c2khjA_
not modelled
26.2
34
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s1;PDBTitle: nmr structure of the domain 6 of the e. coli ribosomal2 protein s1
53 c3iz5Y_
not modelled
25.9
34
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l26 (l24p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
54 c3pifD_
not modelled
25.8
26
PDB header: hydrolaseChain: D: PDB Molecule: 5'->3' exoribonuclease (xrn1);PDBTitle: crystal structure of the 5'->3' exoribonuclease xrn1, e178q mutant in2 complex with manganese
55 c3qnqD_
not modelled
25.6
8
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
56 d1ib8a1
not modelled
25.5
21
Fold: Sm-like foldSuperfamily: YhbC-like, C-terminal domainFamily: YhbC-like, C-terminal domain57 d2pi2e1
not modelled
25.4
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB58 c3bdlA_
not modelled
25.0
23
PDB header: hydrolaseChain: A: PDB Molecule: staphylococcal nuclease domain-containingPDBTitle: crystal structure of a truncated human tudor-sn
59 c3p8bB_
not modelled
24.4
30
PDB header: transferase/transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: x-ray crystal structure of pyrococcus furiosus transcription2 elongation factor spt4/5
60 d1tova_
not modelled
23.9
24
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain61 c1dbgA_
not modelled
23.9
13
PDB header: lyaseChain: A: PDB Molecule: chondroitinase b;PDBTitle: crystal structure of chondroitinase b
62 c4a1cS_
not modelled
23.6
41
PDB header: ribosomeChain: S: PDB Molecule: rpl26;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
63 d2evea1
not modelled
23.4
26
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like64 d1a8pa1
not modelled
22.9
17
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like65 c3hfoC_
not modelled
22.8
14
PDB header: rna binding proteinChain: C: PDB Molecule: ssr3341 protein;PDBTitle: crystal structure of an hfq protein from synechocystis sp.
66 d1sroa_
not modelled
22.8
30
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like67 c2y35A_
not modelled
20.3
23
PDB header: hydrolase/dnaChain: A: PDB Molecule: ld22664p;PDBTitle: crystal structure of xrn1-substrate complex
68 c1vpzB_
not modelled
20.3
21
PDB header: rna binding proteinChain: B: PDB Molecule: carbon storage regulator homolog;PDBTitle: crystal structure of a putative carbon storage regulator protein2 (csra, pa0905) from pseudomonas aeruginosa at 2.05 a resolution
69 c3pr9A_
not modelled
19.0
21
PDB header: chaperoneChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerase;PDBTitle: structural analysis of protein folding by the methanococcus jannaschii2 chaperone fkbp26
70 c2jvvA_
not modelled
18.7
23
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of e. coli nusg carboxyterminal domain
71 c2kvqG_
not modelled
18.7
23
PDB header: transcriptionChain: G: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of nuse:nusg-ctd complex
72 c2jppB_
not modelled
18.6
13
PDB header: translation/rnaChain: B: PDB Molecule: translational repressor;PDBTitle: structural basis of rsma/csra rna recognition: structure of2 rsme bound to the shine-dalgarno sequence of hcna mrna
73 c3qglD_
not modelled
18.5
31
PDB header: protein bindingChain: D: PDB Molecule: sorting nexin-27;PDBTitle: crystal structure of pdz domain of sorting nexin 27 (snx27) in complex2 with the eseskv peptide corresponding to the c-terminal tail of girk3
74 d1vpza_
not modelled
18.3
21
Fold: CsrA-likeSuperfamily: CsrA-likeFamily: CsrA-like75 c2dluA_
not modelled
17.9
26
PDB header: protein bindingChain: A: PDB Molecule: inad-like protein;PDBTitle: solution structure of the second pdz domain of human inad-2 like protein
76 c3ma8A_
not modelled
17.8
27
PDB header: transferaseChain: A: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of cgd1_2040, a pyruvate kinase from cryptosporidium2 parvum
77 d1g31a_
not modelled
17.7
36
Fold: GroES-likeSuperfamily: GroES-likeFamily: GroES78 c2lc4A_
not modelled
17.6
15
PDB header: structural proteinChain: A: PDB Molecule: pilp protein;PDBTitle: solution structure of pilp from pseudomonas aeruginosa
79 d1wfva_
not modelled
17.6
16
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain80 d1nxza1
not modelled
17.4
3
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: YggJ N-terminal domain-like81 d1x5qa1
not modelled
17.4
25
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain82 d1fdra1
not modelled
17.4
25
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like83 d2g2xa1
not modelled
17.3
19
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like84 c3e0vB_
not modelled
17.3
9
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of pyruvate kinase from leishmania mexicana in2 complex with sulphate ions
85 d1fnda1
not modelled
17.1
30
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like86 c2e4hA_
not modelled
17.0
32
PDB header: structural proteinChain: A: PDB Molecule: restin;PDBTitle: solution structure of cytoskeletal protein in complex with2 tubulin tail
87 c2he4A_
not modelled
16.9
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: na(+)/h(+) exchange regulatory cofactor nhe-rf2;PDBTitle: the crystal structure of the second pdz domain of human2 nherf-2 (slc9a3r2) interacting with a mode 1 pdz binding3 motif
88 d2eyqa1
not modelled
16.8
15
Fold: SH3-like barrelSuperfamily: CarD-likeFamily: CarD-like89 d2cp2a1
not modelled
16.6
26
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain90 d2bmwa1
not modelled
16.4
35
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like91 d1nz9a_
not modelled
16.3
26
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain92 c2q9vA_
not modelled
16.1
21
PDB header: transferaseChain: A: PDB Molecule: membrane-associated guanylate kinase, ww and pdz domain-PDBTitle: crystal structure of the c890s mutant of the 4th pdz domain of human2 membrane associated guanylate kinase
93 d1xnea_
not modelled
16.0
26
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain94 d2cp0a1
not modelled
15.8
27
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain95 d1h9ra2
not modelled
15.6
20
Fold: OB-foldSuperfamily: MOP-likeFamily: BiMOP, duplicated molybdate-binding domain96 c1t5aB_
not modelled
15.4
10
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase, m2 isozyme;PDBTitle: human pyruvate kinase m2
97 d2piaa1
not modelled
15.1
24
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like98 c2p5dA_
not modelled
15.1
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0310 protein mjecl36;PDBTitle: crystal structure of mjecl36 from methanocaldococcus2 jannaschii dsm 2661
99 c3iz5G_
not modelled
14.9
23
PDB header: ribosomeChain: G: PDB Molecule: 60s ribosomal protein l6 (l6e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome