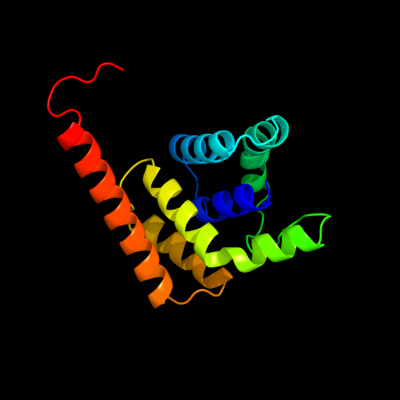
1 d2ou3a1
99.7
17
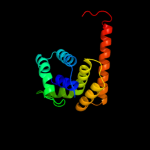
Fold: TerB-likeSuperfamily: TerB-likeFamily: COG3793-like2 c2jxuA_
99.7
15
PDB header: unknown functionChain: A: PDB Molecule: terb;PDBTitle: nmr solution structure of kp-terb, a tellurite resistance2 protein from klebsiella pneumoniae
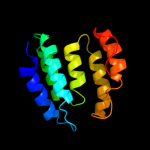
3 c3bvoA_
99.5
14
PDB header: chaperoneChain: A: PDB Molecule: co-chaperone protein hscb, mitochondrial precursor;PDBTitle: crystal structure of human co-chaperone protein hscb
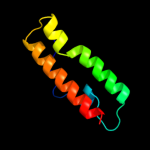
4 c1fpoA_
99.5
22
PDB header: chaperoneChain: A: PDB Molecule: chaperone protein hscb;PDBTitle: hsc20 (hscb), a j-type co-chaperone from e. coli
5 c3hhoA_
99.5
23
PDB header: chaperoneChain: A: PDB Molecule: co-chaperone protein hscb homolog;PDBTitle: chaperone hscb from vibrio cholerae
6 c3apqB_
99.5
31
PDB header: oxidoreductaseChain: B: PDB Molecule: dnaj homolog subfamily c member 10;PDBTitle: crystal structure of j-trx1 fragment of erdj5
7 c2l6lA_
99.5
30
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily c member 24;PDBTitle: solution structure of human j-protein co-chaperone, dph4
8 d1iura_
99.5
20
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain9 d1wjza_
99.4
32
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain10 c2ys8A_
99.4
33
PDB header: protein bindingChain: A: PDB Molecule: rab-related gtp-binding protein rabj;PDBTitle: solution structure of the dnaj-like domain from human ras-2 associated protein rap1
11 c2ctqA_
99.3
28
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily c member 12;PDBTitle: solution structure of j-domain from human dnaj subfamily c2 menber 12
12 d1gh6a_
99.3
22
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain13 c2guzO_
99.3
22
PDB header: chaperone, protein transportChain: O: PDB Molecule: mitochondrial import inner membrane translocasePDBTitle: structure of the tim14-tim16 complex of the mitochondrial2 protein import motor
14 c2yuaA_
99.3
26
PDB header: chaperoneChain: A: PDB Molecule: williams-beuren syndrome chromosome region 18PDBTitle: solution structure of the dnaj domain from human williams-2 beuren syndrome chromosome region 18 protein
15 c2qsaA_
99.3
33
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog dnj-2;PDBTitle: crystal structure of j-domain of dnaj homolog dnj-2 precursor from2 c.elegans.
16 c2dmxA_
99.3
33
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily b member 8;PDBTitle: solution structure of the j domain of dnaj homolog2 subfamily b member 8
17 c3ag7A_
99.3
25
PDB header: plant proteinChain: A: PDB Molecule: putative uncharacterized protein f9e10.5;PDBTitle: an auxilin-like j-domain containing protein, jac1 j-domain
18 d1fpoa1
99.3
22
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain19 c2ctwA_
99.3
30
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily c member 5;PDBTitle: solution structure of j-domain from mouse dnaj subfamily c2 menber 5
20 c2ctrA_
99.3
32
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily b member 9;PDBTitle: solution structure of j-domain from human dnaj subfamily b2 menber 9
21 c2lgwA_
not modelled
99.2
32
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily b member 2;PDBTitle: solution structure of the j domain of hsj1a
22 c2kqxA_
not modelled
99.2
40
PDB header: chaperone binding proteinChain: A: PDB Molecule: curved dna-binding protein;PDBTitle: nmr structure of the j-domain (residues 2-72) in the2 escherichia coli cbpa
23 c2cugA_
not modelled
99.2
38
PDB header: chaperoneChain: A: PDB Molecule: mkiaa0962 protein;PDBTitle: solution structure of the j domain of the pseudo dnaj2 protein, mouse hypothetical mkiaa0962
24 d1fafa_
not modelled
99.2
23
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain25 c2ctpA_
not modelled
99.2
29
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily b member 12;PDBTitle: solution structure of j-domain from human dnaj subfamily b2 menber 12
26 c2dn9A_
not modelled
99.2
30
PDB header: apoptosis, chaperoneChain: A: PDB Molecule: dnaj homolog subfamily a member 3;PDBTitle: solution structure of j-domain from the dnaj homolog, human2 tid1 protein
27 d1n4ca_
not modelled
99.2
27
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain28 d1nz6a_
not modelled
99.1
26
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain29 c2pf4E_
not modelled
99.1
21
PDB header: hydrolase regulator/viral proteinChain: E: PDB Molecule: small t antigen;PDBTitle: crystal structure of the full-length simian virus 40 small t antigen2 complexed with the protein phosphatase 2a aalpha subunit
30 d1hdja_
not modelled
99.1
35
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain31 d1xbla_
not modelled
99.1
33
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain32 c2o37A_
not modelled
99.1
31
PDB header: chaperoneChain: A: PDB Molecule: protein sis1;PDBTitle: j-domain of sis1 protein, hsp40 co-chaperone from2 saccharomyces cerevisiae.
33 c2ochA_
not modelled
99.1
38
PDB header: chaperoneChain: A: PDB Molecule: hypothetical protein dnj-12;PDBTitle: j-domain of dnj-12 from caenorhabditis elegans
34 c2guzD_
not modelled
99.0
18
PDB header: chaperone, protein transportChain: D: PDB Molecule: mitochondrial import inner membrane translocasePDBTitle: structure of the tim14-tim16 complex of the mitochondrial2 protein import motor
35 c1bq0A_
not modelled
99.0
34
PDB header: chaperoneChain: A: PDB Molecule: dnaj;PDBTitle: j-domain (residues 1-77) of the escherichia coli n-terminal2 fragment (residues 1-104) of the molecular chaperone dnaj,3 nmr, 20 structures
36 c2y4tA_
not modelled
98.8
16
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily c member 3;PDBTitle: crystal structure of the human co-chaperone p58(ipk)
37 c3uo2A_
not modelled
97.5
23
PDB header: chaperoneChain: A: PDB Molecule: j-type co-chaperone jac1, mitochondrial;PDBTitle: jac1 co-chaperone from saccharomyces cerevisiae
38 d2h5na1
not modelled
97.2
11
Fold: TerB-likeSuperfamily: TerB-likeFamily: PG1108-like39 c3apoA_
not modelled
95.2
32
PDB header: oxidoreductaseChain: A: PDB Molecule: dnaj homolog subfamily c member 10;PDBTitle: crystal structure of full-length erdj5
40 d1ug2a_
not modelled
92.3
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain41 c3evrA_
not modelled
77.9
10
PDB header: signaling proteinChain: A: PDB Molecule: myosin light chain kinase, green fluorescentPDBTitle: crystal structure of calcium bound monomeric gcamp2
42 d2ouwa1
not modelled
75.9
20
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: TTHA0727-like43 d1aisb2
not modelled
73.1
17
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Transcription factor IIB (TFIIB), core domain44 c2hpkA_
not modelled
69.3
13
PDB header: luminescent proteinChain: A: PDB Molecule: photoprotein berovin;PDBTitle: crystal structure of photoprotein berovin from beroe2 abyssicola
45 d2jxca1
not modelled
67.1
7
Fold: EF Hand-likeSuperfamily: EF-handFamily: Eps15 homology domain (EH domain)46 d1vola2
not modelled
62.7
12
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Transcription factor IIB (TFIIB), core domain47 c2iqcA_
59.6
15
PDB header: protein bindingChain: A: PDB Molecule: fanconi anemia group f protein;PDBTitle: crystal structure of human fancf protein that functions in2 the assembly of a dna damage signaling complex
48 c2aucC_
not modelled
59.5
13
PDB header: membrane proteinChain: C: PDB Molecule: myosin a tail interacting protein;PDBTitle: structure of the plasmodium mtip-myoa complex, a key component of the2 parasite invasion motor
49 d1vola1
not modelled
53.9
12
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Transcription factor IIB (TFIIB), core domain50 d1a04a1
not modelled
49.6
30
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)51 c2q0oA_
not modelled
46.5
23
PDB header: transcriptionChain: A: PDB Molecule: probable transcriptional activator protein trar;PDBTitle: crystal structure of an anti-activation complex in bacterial quorum2 sensing
52 c3k7aM_
not modelled
44.5
5
PDB header: transcriptionChain: M: PDB Molecule: transcription initiation factor iib;PDBTitle: crystal structure of an rna polymerase ii-tfiib complex
53 c2kxaA_
not modelled
44.0
32
PDB header: viral protein, immune systemChain: A: PDB Molecule: haemagglutinin ha2 chain peptide;PDBTitle: the hemagglutinin fusion peptide (h1 subtype) at ph 7.4
54 c1h0mD_
not modelled
42.2
27
PDB header: transcription/dnaChain: D: PDB Molecule: transcriptional activator protein trar;PDBTitle: three-dimensional structure of the quorum sensing protein2 trar bound to its autoinducer and to its target dna
55 c2ggzB_
not modelled
41.2
13
PDB header: lyase activatorChain: B: PDB Molecule: guanylyl cyclase-activating protein 3;PDBTitle: crystal structure of human guanylate cyclase activating2 protein-3
56 c2kfhA_
not modelled
39.7
18
PDB header: protein bindingChain: A: PDB Molecule: eh domain-containing protein 1;PDBTitle: structure of the c-terminal domain of ehd1 with fnyestgpftak
57 d1aisb1
not modelled
39.4
18
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Transcription factor IIB (TFIIB), core domain58 c2in3A_
not modelled
38.9
14
PDB header: isomeraseChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a putative protein disulfide isomerase from2 nitrosomonas europaea
59 c3hugJ_
not modelled
38.7
25
PDB header: transcription/membrane proteinChain: J: PDB Molecule: probable conserved membrane protein;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
60 c1yzxB_
not modelled
38.5
13
PDB header: transferaseChain: B: PDB Molecule: glutathione s-transferase kappa 1;PDBTitle: crystal structure of human kappa class glutathione2 transferase
61 d1fi6a_
not modelled
37.1
16
Fold: EF Hand-likeSuperfamily: EF-handFamily: Eps15 homology domain (EH domain)62 c3fiaA_
not modelled
37.0
9
PDB header: protein bindingChain: A: PDB Molecule: intersectin-1;PDBTitle: crystal structure of the eh 1 domain from human intersectin-2 1 protein. northeast structural genomics consortium target3 hr3646e.
63 d1rioa_
not modelled
36.6
15
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors64 d1c07a_
not modelled
36.5
7
Fold: EF Hand-likeSuperfamily: EF-handFamily: Eps15 homology domain (EH domain)65 c3kzqE_
not modelled
35.1
19
PDB header: structural genomics, unknown functionChain: E: PDB Molecule: putative uncharacterized protein vp2116;PDBTitle: the crystal structure of the protein with unknown function from vibrio2 parahaemolyticus rimd 2210633
66 d1vkea_
not modelled
34.4
17
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: CMD-like67 c3oouA_
not modelled
34.3
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin2118 protein;PDBTitle: the structure of a protein with unkown function from listeria innocua
68 d1p4wa_
not modelled
34.2
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)69 d1r4wa_
not modelled
34.2
14
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbA-like70 d2mysb_
not modelled
34.0
10
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like71 c2pmyB_
not modelled
33.9
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ras and ef-hand domain-containing protein;PDBTitle: ef-hand domain of human rasef
72 d1v4wa_
not modelled
33.1
9
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins73 c3ek7A_
not modelled
32.2
13
PDB header: fluorescent proteinChain: A: PDB Molecule: myosin light chain kinase, green fluorescentPDBTitle: calcium-saturated gcamp2 dimer
74 c1w8jD_
not modelled
32.0
19
PDB header: motor proteinChain: D: PDB Molecule: myosin va;PDBTitle: crystal structure of myosin v motor domain -2 nucleotide-free
75 c2hg5D_
not modelled
32.0
24
PDB header: membrane proteinChain: D: PDB Molecule: kcsa channel;PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
76 c1o17A_
not modelled
30.9
16
PDB header: transferaseChain: A: PDB Molecule: anthranilate phosphoribosyltransferase;PDBTitle: anthranilate phosphoribosyl-transferase (trpd)
77 c3mseB_
not modelled
30.4
11
PDB header: transferaseChain: B: PDB Molecule: calcium-dependent protein kinase, putative;PDBTitle: crystal structure of c-terminal domain of pf110239.
78 c3ivpD_
not modelled
30.0
17
PDB header: dna binding proteinChain: D: PDB Molecule: putative transposon-related dna-binding protein;PDBTitle: the structure of a possible transposon-related dna-binding protein2 from clostridium difficile 630.
79 d3e11a1
not modelled
29.1
26
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: TTHA0227-like80 c3j04A_
not modelled
29.1
10
PDB header: structural proteinChain: A: PDB Molecule: myosin-11;PDBTitle: em structure of the heavy meromyosin subfragment of chick smooth2 muscle myosin with regulatory light chain in phosphorylated state
81 c2doqA_
not modelled
29.0
7
PDB header: cell cycleChain: A: PDB Molecule: cell division control protein 31;PDBTitle: crystal structure of sfi1p/cdc31p complex
82 c3t76A_
not modelled
28.9
20
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator vanug;PDBTitle: crystal structure of transcriptional regulator vanug, form ii
83 c2rmrA_
not modelled
28.7
15
PDB header: transcriptionChain: A: PDB Molecule: paired amphipathic helix protein sin3a;PDBTitle: solution structure of msin3a pah1 domain
84 c2otgA_
not modelled
28.3
11
PDB header: contractile proteinChain: A: PDB Molecule: myosin heavy chain;PDBTitle: rigor-like structures of muscle myosins reveal key2 mechanical elements in the transduction pathways of this3 allosteric motor
85 c1p8cD_
not modelled
28.2
16
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of tm1620 (apc4843) from thermotoga2 maritima
86 c1eg4A_
not modelled
27.0
14
PDB header: structural proteinChain: A: PDB Molecule: dystrophin;PDBTitle: structure of a dystrophin ww domain fragment in complex2 with a beta-dystroglycan peptide
87 d1qjta_
not modelled
26.7
18
Fold: EF Hand-likeSuperfamily: EF-handFamily: Eps15 homology domain (EH domain)88 c3eusB_
not modelled
25.7
10
PDB header: dna binding proteinChain: B: PDB Molecule: dna-binding protein;PDBTitle: the crystal structure of the dna binding protein from silicibacter2 pomeroyi
89 c2bl0C_
not modelled
25.3
9
PDB header: muscle proteinChain: C: PDB Molecule: myosin regulatory light chain;PDBTitle: physarum polycephalum myosin ii regulatory domain
90 c2kvcA_
not modelled
25.1
7
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: solution structure of the mycobacterium tuberculosis protein rv0543c,2 a member of the duf3349 superfamily. seattle structural genomics3 center for infectious disease target mytud.17112.a
91 d1sq8a_
not modelled
25.1
41
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors92 d1ha7a_
not modelled
24.8
12
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins93 d1b33b_
not modelled
24.6
14
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins94 c2kgrA_
not modelled
24.6
12
PDB header: protein bindingChain: A: PDB Molecule: intersectin-1;PDBTitle: solution structure of protein itsn1 from homo sapiens.2 northeast structural genomics consortium target hr5524a
95 d1allb_
not modelled
24.4
15
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins96 c2dfsA_
not modelled
24.4
19
PDB header: contractile protein/transport proteinChain: A: PDB Molecule: myosin-5a;PDBTitle: 3-d structure of myosin-v inhibited state
97 c3f6wE_
not modelled
23.9
16
PDB header: dna binding proteinChain: E: PDB Molecule: xre-family like protein;PDBTitle: xre-family like protein from pseudomonas syringae pv. tomato str.2 dc3000
98 c1i84V_
not modelled
22.9
10
PDB header: contractile proteinChain: V: PDB Molecule: smooth muscle myosin heavy chain;PDBTitle: cryo-em structure of the heavy meromyosin subfragment of2 chicken gizzard smooth muscle myosin with regulatory light3 chain in the dephosphorylated state. only c alphas4 provided for regulatory light chain. only backbone atoms5 provided for s2 fragment.
99 d2croa_
not modelled
22.8
34
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors100 d1jeba_
not modelled
22.2
11
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins101 c3d7iB_
not modelled
21.7
21
PDB header: lyaseChain: B: PDB Molecule: carboxymuconolactone decarboxylase family protein;PDBTitle: crystal structure of carboxymuconolactone decarboxylase family protein2 possibly involved in oxygen detoxification (1591455) from3 methanococcus jannaschii at 1.75 a resolution
102 c3k69A_
not modelled
21.6
15
PDB header: transcriptionChain: A: PDB Molecule: putative transcription regulator;PDBTitle: crystal structure of a putative transcriptional regulator (lp_0360)2 from lactobacillus plantarum at 1.95 a resolution
103 d2gqba1
not modelled
21.6
31
Fold: RPA2825-likeSuperfamily: RPA2825-likeFamily: RPA2825-like104 c3op9A_
not modelled
21.5
11
PDB header: transcription regulatorChain: A: PDB Molecule: pli0006 protein;PDBTitle: crystal structure of transcriptional regulator from listeria innocua
105 c3trbA_
not modelled
21.4
19
PDB header: dna binding proteinChain: A: PDB Molecule: virulence-associated protein i;PDBTitle: structure of an addiction module antidote protein of a higa (higa)2 family from coxiella burnetii
106 d1wdcb_
not modelled
21.4
8
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like107 d1ffva1
not modelled
21.3
21
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like108 c3sibA_
not modelled
21.3
11
PDB header: dna binding proteinChain: A: PDB Molecule: ure3-bp sequence specific dna binding protein;PDBTitle: crystal structure of ure3-binding protein, wild-type
109 c2gbbA_
not modelled
21.2
16
PDB header: isomeraseChain: A: PDB Molecule: putative chorismate mutase;PDBTitle: crystal structure of secreted chorismate mutase from2 yersinia pestis
110 d1n62a1
not modelled
20.7
18
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like111 c2ebyA_
not modelled
20.7
14
PDB header: transcriptionChain: A: PDB Molecule: putative hth-type transcriptional regulator ybaq;PDBTitle: crystal structure of a hypothetical protein from e. coli
112 c1br4E_
not modelled
20.2
10
PDB header: muscle proteinChain: E: PDB Molecule: myosin;PDBTitle: smooth muscle myosin motor domain-essential light chain2 complex with mgadp.bef3 bound at the active site
113 d1b8da_
not modelled
20.1
8
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins114 d1f99a_
not modelled
20.0
12
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins