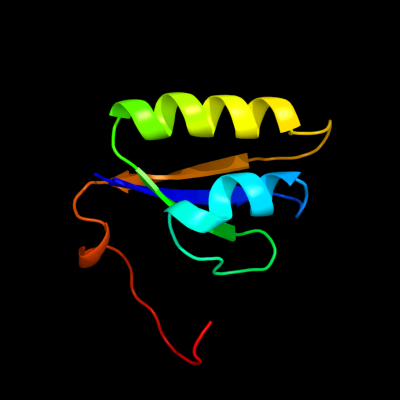
1 c3oq2A_
71.3
14
PDB header: immune systemChain: A: PDB Molecule: crispr-associated protein cas2;PDBTitle: structure of a crispr associated protein cas2 from desulfovibrio2 vulgaris
2 d1zpwx1
56.3
17
Fold: Ferredoxin-likeSuperfamily: TTP0101/SSO1404-likeFamily: TTP0101/SSO1404-like3 d2i0xa1
44.4
24
Fold: Ferredoxin-likeSuperfamily: TTP0101/SSO1404-likeFamily: TTP0101/SSO1404-like4 d2ivya1
39.2
16
Fold: Ferredoxin-likeSuperfamily: TTP0101/SSO1404-likeFamily: TTP0101/SSO1404-like5 c1yz4A_
34.1
20
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase-like 15 isoform a;PDBTitle: crystal structure of dusp15
6 c2p4dA_
29.1
26
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase;PDBTitle: structure-assisted discovery of variola major h12 phosphatase inhibitors
7 d1m3ga_
22.5
12
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like8 c2e5jA_
22.4
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: methenyltetrahydrofolate synthetase domainPDBTitle: solution structure of rna binding domain in2 methenyltetrahydrofolate synthetase domain containing
9 c1zzwA_
21.6
13
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 10;PDBTitle: crystal structure of catalytic domain of human map kinase2 phosphatase 5
10 d1h4vb1
20.8
13
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS11 d1wf0a_
20.5
23
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD12 d1x52a1
20.4
10
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: ERF1/Dom34 C-terminal domain-like13 c3cwvB_
20.0
20
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase, b subunit, truncated;PDBTitle: crystal structure of b-subunit of the dna gyrase from myxococcus2 xanthus
14 c2dgyA_
19.8
19
PDB header: translationChain: A: PDB Molecule: mgc11102 protein;PDBTitle: solution structure of the eukaryotic initiation factor 1a2 in mgc11102 protein
15 d1f20a2
19.5
8
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: NADPH-cytochrome p450 reductase-like16 c2r0bA_
19.0
13
PDB header: hydrolaseChain: A: PDB Molecule: serine/threonine/tyrosine-interacting protein;PDBTitle: crystal structure of human tyrosine phosphatase-like2 serine/threonine/tyrosine-interacting protein
17 c2oqkA_
16.3
14
PDB header: translationChain: A: PDB Molecule: putative translation initiation factor eif-1a;PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
18 c3lnuA_
15.7
30
PDB header: isomeraseChain: A: PDB Molecule: topoisomerase iv subunit b;PDBTitle: crystal structure of pare subunit
19 c1no8A_
15.7
13
PDB header: rna binding proteinChain: A: PDB Molecule: aly;PDBTitle: solution structure of the nuclear factor aly rbd domain
20 d1no8a_
15.7
13
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD21 c2osrA_
not modelled
14.7
9
PDB header: rna binding proteinChain: A: PDB Molecule: nucleolar protein 3;PDBTitle: nmr structure of rrm-2 of yeast npl3 protein
22 c2esbA_
not modelled
14.6
20
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 18;PDBTitle: crystal structure of human dusp18
23 c3d0jA_
not modelled
14.3
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ca_c3497;PDBTitle: crystal structure of conserved protein of unknown function ca_c34972 from clostridium acetobutylicum atcc 824
24 c2e0tA_
not modelled
14.3
35
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase 26;PDBTitle: crystal structure of catalytic domain of dual specificity phosphatase2 26, ms0830 from homo sapiens
25 c3pgwP_
not modelled
14.3
14
PDB header: splicing/dna/rnaChain: P: PDB Molecule: u1-a;PDBTitle: crystal structure of human u1 snrnp
26 c2wgpA_
not modelled
14.2
8
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 14;PDBTitle: crystal structure of human dual specificity phosphatase 14
27 c2y96A_
not modelled
13.4
36
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase dupd1;PDBTitle: structure of human dual-specificity phosphatase 27
28 c2oudA_
not modelled
13.2
14
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 10;PDBTitle: crystal structure of the catalytic domain of human mkp5
29 c2hcmA_
not modelled
13.2
17
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase;PDBTitle: crystal structure of mouse putative dual specificity phosphatase2 complexed with zinc tungstate, new york structural genomics3 consortium
30 c1wrmA_
not modelled
12.9
20
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase 22;PDBTitle: crystal structure of jsp-1
31 c3ek1C_
not modelled
12.6
20
PDB header: oxidoreductaseChain: C: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from brucella2 melitensis biovar abortus 2308
32 d1jl0a_
not modelled
12.4
12
Fold: S-adenosylmethionine decarboxylaseSuperfamily: S-adenosylmethionine decarboxylaseFamily: S-adenosylmethionine decarboxylase33 c3jz4C_
not modelled
11.8
25
PDB header: oxidoreductaseChain: C: PDB Molecule: succinate-semialdehyde dehydrogenase [nadp+];PDBTitle: crystal structure of e. coli nadp dependent enzyme
34 d2f9da1
not modelled
10.9
18
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD35 d1pvga2
not modelled
10.6
24
Fold: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseSuperfamily: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseFamily: DNA gyrase/MutL, N-terminal domain36 c1qzrA_
not modelled
10.1
24
PDB header: isomeraseChain: A: PDB Molecule: dna topoisomerase ii;PDBTitle: crystal structure of the atpase region of saccharomyces cerevisiae2 topoisomerase ii bound to icrf-187 (dexrazoxane)
37 d2vgna3
not modelled
10.1
15
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: ERF1/Dom34 C-terminal domain-like38 c3emuA_
not modelled
10.0
14
PDB header: hydrolaseChain: A: PDB Molecule: leucine rich repeat and phosphatase domainPDBTitle: crystal structure of a leucine rich repeat and phosphatase2 domain containing protein from entamoeba histolytica
39 d1mkpa_
not modelled
9.9
21
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like40 d2ghpa2
not modelled
9.8
13
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD41 d1vhra_
not modelled
9.6
38
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like42 c1zxnB_
not modelled
9.4
24
PDB header: isomeraseChain: B: PDB Molecule: dna topoisomerase ii, alpha isozyme;PDBTitle: human dna topoisomerase iia atpase/adp
43 d1uawa_
not modelled
9.2
19
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD44 c2kkuA_
not modelled
9.0
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of protein af2351 from archaeoglobus2 fulgidus. northeast structural genomics consortium target3 att9/ontario center for structural proteomics target af2351
45 d1o04a_
not modelled
8.8
18
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like46 c3pgwS_
not modelled
8.7
12
PDB header: splicing/dna/rnaChain: S: PDB Molecule: u1-70k;PDBTitle: crystal structure of human u1 snrnp
47 c2g6zB_
not modelled
8.5
27
PDB header: hydrolaseChain: B: PDB Molecule: dual specificity protein phosphatase 5;PDBTitle: crystal structure of human dusp5
48 c1flcB_
not modelled
8.5
25
PDB header: hydrolaseChain: B: PDB Molecule: haemagglutinin-esterase-fusion glycoprotein;PDBTitle: x-ray structure of the haemagglutinin-esterase-fusion glycoprotein of2 influenza c virus
49 c2e44A_
not modelled
8.3
9
PDB header: rna binding proteinChain: A: PDB Molecule: insulin-like growth factor 2 mrna bindingPDBTitle: solution structure of rna binding domain in insulin-like2 growth factor 2 mrna binding protein 3
50 d2cq3a1
not modelled
8.2
20
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD51 d1sqwa1
not modelled
8.0
14
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain52 d1vpqa_
not modelled
8.0
19
Fold: TIM beta/alpha-barrelSuperfamily: TM1631-likeFamily: TM1631-like53 c2gwoC_
not modelled
7.9
33
PDB header: hydrolaseChain: C: PDB Molecule: dual specificity protein phosphatase 13;PDBTitle: crystal structure of tmdp
54 c3ifgH_
not modelled
7.6
21
PDB header: oxidoreductaseChain: H: PDB Molecule: succinate-semialdehyde dehydrogenase (nadp+);PDBTitle: crystal structure of succinate-semialdehyde dehydrogenase from2 burkholderia pseudomallei, part 1 of 2
55 d2cpja1
not modelled
7.6
23
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD56 c3lyrA_
not modelled
7.6
13
PDB header: transcription activatorChain: A: PDB Molecule: transcription factor coe1;PDBTitle: human early b-cell factor 1 (ebf1) dna-binding domain
57 c1s16B_
not modelled
7.4
26
PDB header: isomeraseChain: B: PDB Molecule: topoisomerase iv subunit b;PDBTitle: crystal structure of e. coli topoisomerase iv pare 43kda subunit2 complexed with adpnp
58 c2nt2C_
not modelled
7.3
18
PDB header: hydrolaseChain: C: PDB Molecule: protein phosphatase slingshot homolog 2;PDBTitle: crystal structure of slingshot phosphatase 2
59 c2vgmA_
not modelled
7.2
15
PDB header: cell cycleChain: A: PDB Molecule: dom34;PDBTitle: structure of yeast dom34 : a protein related to translation2 termination factor erf1 and involved in no-go decay.
60 c3ed6B_
not modelled
6.9
16
PDB header: oxidoreductaseChain: B: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: 1.7 angstrom resolution crystal structure of betaine aldehyde2 dehydrogenase (betb) from staphylococcus aureus
61 c3b4wA_
not modelled
6.8
16
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of mycobacterium tuberculosis aldehyde dehydrogenase2 complexed with nad+
62 c3s01A_
not modelled
6.8
11
PDB header: rna binding proteinChain: A: PDB Molecule: heterogeneous nuclear ribonucleoprotein l;PDBTitle: crystal structure of a heterogeneous nuclear ribonucleoprotein l2 (hnrpl) from mus musculus at 2.15 a resolution
63 c3bs9A_
not modelled
6.8
19
PDB header: rna binding proteinChain: A: PDB Molecule: nucleolysin tia-1 isoform p40;PDBTitle: x-ray structure of human tia-1 rrm2
64 d1fjca_
not modelled
6.7
14
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD65 c2hvzA_
not modelled
6.7
23
PDB header: rna binding proteinChain: A: PDB Molecule: splicing factor, arginine/serine-rich 7;PDBTitle: solution structure of the rrm domain of sr rich factor 9g8
66 c3p6yD_
not modelled
6.7
7
PDB header: rna binding protein/rnaChain: D: PDB Molecule: cleavage and polyadenylation specificity factor subunit 6;PDBTitle: cf im25-cf im68-uguaa complex
67 d1y0na_
not modelled
6.6
14
Fold: YehU-likeSuperfamily: YehU-likeFamily: YehU-like68 d1x4aa1
not modelled
6.5
19
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD69 d1fnxh1
not modelled
6.5
10
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD70 c2fhoB_
not modelled
6.4
17
PDB header: rna binding proteinChain: B: PDB Molecule: spliceosomal protein p14;PDBTitle: nmr solution structure of the human spliceosomal protein2 complex p14-sf3b155
71 c2i38A_
not modelled
6.3
23
PDB header: rna binding protein/chimeraChain: A: PDB Molecule: fusion protein consists of immunoglobin g-PDBTitle: solution structure of the rrm of srp20
72 c3mlpE_
not modelled
6.2
13
PDB header: transcription/dnaChain: E: PDB Molecule: transcription factor coe1;PDBTitle: early b-cell factor 1 (ebf1) bound to dna
73 c2zamA_
not modelled
6.1
21
PDB header: protein transportChain: A: PDB Molecule: vacuolar protein sorting-associating protein 4b;PDBTitle: crystal structure of mouse skd1/vps4b apo-form
74 c3r31A_
not modelled
6.1
20
PDB header: oxidoreductaseChain: A: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: crystal structure of betaine aldehyde dehydrogenase from agrobacterium2 tumefaciens
75 c2e5gA_
not modelled
6.0
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: u6 snrna-specific terminal uridylyltransferase 1;PDBTitle: solution structure of rna binding domain in rna binding2 motif protein 21
76 d2ghpa3
not modelled
6.0
11
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD77 d2qwva1
not modelled
5.9
29
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: AF1056-like78 c2krbA_
not modelled
5.8
15
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 3PDBTitle: solution structure of eif3b-rrm bound to eif3j peptide
79 d2cqia1
not modelled
5.8
17
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD80 c1ei1B_
not modelled
5.8
28
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase b;PDBTitle: dimerization of e. coli dna gyrase b provides a structural mechanism2 for activating the atpase catalytic center
81 c2vroB_
not modelled
5.7
27
PDB header: oxidoreductaseChain: B: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from2 burkholderia xenovorans lb400
82 c2dngA_
not modelled
5.7
20
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 4h;PDBTitle: solution structure of rna binding domain in eukaryotic2 translation initiation factor 4h
83 c1t90B_
not modelled
5.6
20
PDB header: oxidoreductaseChain: B: PDB Molecule: probable methylmalonate-semialdehydePDBTitle: crystal structure of methylmalonate semialdehyde2 dehydrogenase from bacillus subtilis
84 d1bxsa_
not modelled
5.6
19
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like85 c3ep3A_
not modelled
5.6
17
PDB header: lyaseChain: A: PDB Molecule: s-adenosylmethionine decarboxylase alpha chain;PDBTitle: human adometdc d174n mutant with no putrescine bound
86 c3mcaB_
not modelled
5.5
14
PDB header: translation regulation/hydrolaseChain: B: PDB Molecule: protein dom34;PDBTitle: structure of the dom34-hbs1 complex and implications for its role in2 no-go decay
87 d1fjeb2
not modelled
5.5
14
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD88 d1wi8a_
not modelled
5.5
12
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD89 d1a4sa_
not modelled
5.4
20
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like90 d1ynya1
not modelled
5.3
15
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: Hydantoinase (dihydropyrimidinase)91 d1o89a2
not modelled
5.3
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain92 d3dhwc1
not modelled
5.3
9
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like93 d1x0fa1
not modelled
5.2
28
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD94 c3cp0A_
not modelled
5.2
13
PDB header: membrane proteinChain: A: PDB Molecule: membrane protein implicated in regulation of membranePDBTitle: crystal structure of the soluble domain of membrane protein implicated2 in regulation of membrane protease activity from corynebacterium3 glutamicum
95 d1ky8a_
not modelled
5.2
21
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like96 c2fc9A_
not modelled
5.2
14
PDB header: rna binding proteinChain: A: PDB Molecule: ncl protein;PDBTitle: solution structure of the rrm_1 domain of ncl protein
97 c2fc8A_
not modelled
5.1
18
PDB header: rna binding proteinChain: A: PDB Molecule: ncl protein;PDBTitle: solution structure of the rrm_1 domain of ncl protein