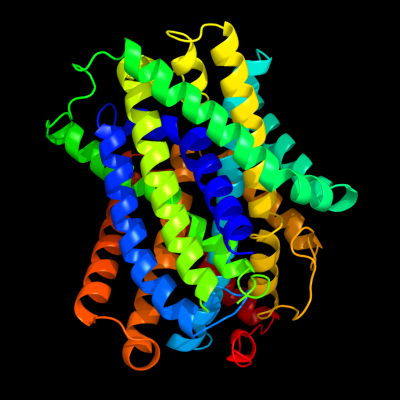
| 1 | c3giaA_
|
|
 |
97.0 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
|
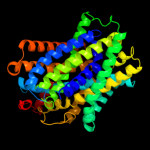
| 2 | c2jlnA_
|
|
 |
96.9 |
15 |
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
|
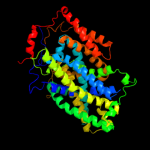
| 3 | c3lrcC_
|
|
 |
94.8 |
15 |
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
|
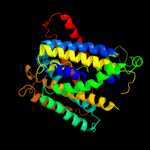
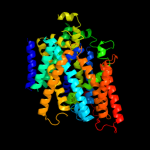
| 4 | c2xq2A_
|
|
 |
94.0 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
|
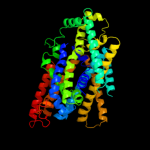
| 5 | c3dh4A_
|
|
 |
87.2 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
|
| 6 | c3a8qB_
|
|
 |
42.1 |
70 |
PDB header:signaling protein
Chain: B: PDB Molecule:t-lymphoma invasion and metastasis-inducing
PDBTitle: low-resolution crystal structure of the tiam2 phccex domain
|
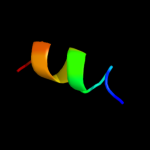
| 7 | c3a8nA_
|
|
 |
40.7 |
58 |
PDB header:signaling protein
Chain: A: PDB Molecule:t-lymphoma invasion and metastasis-inducing
PDBTitle: crystal structure of the tiam1 phccex domain
|
| 8 | c3pl0B_
|
|
 |
38.2 |
44 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a bsma homolog (mpe_a2762) from methylobium2 petroleophilum pm1 at 1.91 a resolution
|
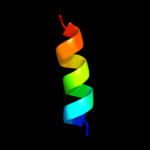
| 9 | d2axtj1
|
|
 |
27.7 |
36 |
Fold:Single transmembrane helix
Superfamily:Photosystem II reaction center protein J, PsbJ
Family:PsbJ-like |
| 10 | d2bfdb2
|
|
 |
27.3 |
4 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
| 11 | d1rp4a_
|
|
 |
26.8 |
36 |
Fold:ERO1-like
Superfamily:ERO1-like
Family:ERO1-like |
| 12 | c3hd6A_
|
|
 |
26.3 |
14 |
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
|
| 13 | c2k9pA_
|
|
 |
26.3 |
18 |
PDB header:membrane protein
Chain: A: PDB Molecule:pheromone alpha factor receptor;
PDBTitle: structure of tm1_tm2 in lppg micelles
|
| 14 | d1h5pa_
|
|
 |
24.8 |
33 |
Fold:SAND domain-like
Superfamily:SAND domain-like
Family:SAND domain |
| 15 | c2ktlA_
|
|
 |
24.5 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:tyrosyl-trna synthetase;
PDBTitle: structure of c-terminal domain from mttyrrs of a. nidulans
|
| 16 | d1qs0b2
|
|
 |
24.1 |
15 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
| 17 | d2jeka1
|
|
 |
23.7 |
19 |
Fold:Rv1873-like
Superfamily:Rv1873-like
Family:Rv1873-like |
| 18 | d1oqja_
|
|
 |
23.6 |
20 |
Fold:SAND domain-like
Superfamily:SAND domain-like
Family:SAND domain |
| 19 | d1s6la1
|
|
 |
23.5 |
31 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MerB N-terminal domain-like |
| 20 | c3b9yA_
|
|
 |
23.3 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
|
| 21 | d1afra_ |
|
not modelled |
23.0 |
30 |
Fold:Ferritin-like
Superfamily:Ferritin-like
Family:Ribonucleotide reductase-like |
| 22 | d1ufna_ |
|
not modelled |
22.4 |
27 |
Fold:SAND domain-like
Superfamily:SAND domain-like
Family:SAND domain |
| 23 | c3zy6A_ |
|
not modelled |
22.4 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:putative gdp-fucose protein o-fucosyltransferase 1;
PDBTitle: crystal structure of pofut1 in complex with gdp-fucose2 (crystal-form-ii)
|
| 24 | d1ik6a2 |
|
not modelled |
21.2 |
12 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
| 25 | d2awia1 |
|
not modelled |
20.7 |
29 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:PrgX N-terminal domain-like |
| 26 | c3iymA_ |
|
not modelled |
20.6 |
35 |
PDB header:virus
Chain: A: PDB Molecule:capsid protein;
PDBTitle: backbone trace of the capsid protein dimer of a fungal partitivirus2 from electron cryomicroscopy and homology modeling
|
| 27 | c2qx5B_ |
|
not modelled |
19.9 |
12 |
PDB header:transport protein
Chain: B: PDB Molecule:nucleoporin nic96;
PDBTitle: structure of nucleoporin nic96
|
| 28 | d2ozlb2 |
|
not modelled |
19.5 |
10 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
| 29 | c3ahrA_ |
|
not modelled |
19.0 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ero1-like protein alpha;
PDBTitle: inactive human ero1
|
| 30 | c3pppA_ |
|
not modelled |
18.8 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:glycine betaine/carnitine/choline-binding protein;
PDBTitle: structures of the substrate-binding protein provide insights into the2 multiple compatible solutes binding specificities of bacillus3 subtilis abc transporter opuc
|
| 31 | d1w85b2 |
|
not modelled |
17.5 |
28 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
| 32 | d2fug61 |
|
not modelled |
17.3 |
36 |
Fold:HydA/Nqo6-like
Superfamily:HydA/Nqo6-like
Family:Nq06-like |
| 33 | d1i0za2 |
|
not modelled |
17.2 |
21 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 34 | d1llda2 |
|
not modelled |
17.0 |
29 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 35 | d2yrka1 |
|
not modelled |
16.8 |
27 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:HkH motif-containing C2H2 finger |
| 36 | d9ldta2 |
|
not modelled |
16.7 |
25 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 37 | c2lbfA_ |
|
not modelled |
16.6 |
43 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:60s acidic ribosomal protein p1;
PDBTitle: solution structure of the dimerization domain of human ribosomal2 protein p1/p2 heterodimer
|
| 38 | d1nekd_ |
|
not modelled |
16.6 |
13 |
Fold:Heme-binding four-helical bundle
Superfamily:Fumarate reductase respiratory complex transmembrane subunits
Family:Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD) |
| 39 | c2k42A_ |
|
not modelled |
16.4 |
15 |
PDB header:signaling protein
Chain: A: PDB Molecule:wiskott-aldrich syndrome protein;
PDBTitle: solution structure of the gtpase binding domain of wasp in2 complex with espfu, an ehec effector
|
| 40 | c2elnA_ |
|
not modelled |
16.2 |
50 |
PDB header:transcription
Chain: A: PDB Molecule:zinc finger protein 406;
PDBTitle: solution structure of the 11th c2h2 zinc finger of human2 zinc finger protein 406
|
| 41 | c3iz5t_ |
|
not modelled |
15.8 |
43 |
PDB header:ribosome
Chain: T: PDB Molecule:60s ribosomal protein l19 (l19e);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 42 | c3nctC_ |
|
not modelled |
15.8 |
22 |
PDB header:dna binding protein, chaperone
Chain: C: PDB Molecule:protein psib;
PDBTitle: x-ray crystal structure of the bacterial conjugation factor psib, a2 negative regulator of reca
|
| 43 | c2etjA_ |
|
not modelled |
15.7 |
67 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease hii;
PDBTitle: crystal structure of ribonuclease hii (ec 3.1.26.4) (rnase hii)2 (tm0915) from thermotoga maritima at 1.74 a resolution
|
| 44 | d2etja1 |
|
not modelled |
15.7 |
67 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 45 | d1fvka_ |
|
not modelled |
15.7 |
29 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:DsbA-like |
| 46 | d1i10a2 |
|
not modelled |
15.7 |
25 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 47 | d1umdb2 |
|
not modelled |
15.6 |
19 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
| 48 | c1x4rA_ |
|
not modelled |
15.2 |
29 |
PDB header:apoptosis
Chain: A: PDB Molecule:parp14 protein;
PDBTitle: solution structure of wwe domain in parp14 protein
|
| 49 | d1oeya_ |
|
not modelled |
15.0 |
20 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:CAD & PB1 domains
Family:PB1 domain |
| 50 | d1d4va1 |
|
not modelled |
14.9 |
38 |
Fold:TNF receptor-like
Superfamily:TNF receptor-like
Family:TNF receptor-like |
| 51 | c2k1aA_ |
|
not modelled |
14.8 |
45 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
|
| 52 | c3es4B_ |
|
not modelled |
14.2 |
31 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein duf861 with a rmlc-like cupin fold;
PDBTitle: crystal structure of protein of unknown function (duf861) with a rmlc-2 like cupin fold (17741406) from agrobacterium tumefaciens str. c583 (dupont) at 1.64 a resolution
|
| 53 | c3o66A_ |
|
not modelled |
14.2 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:glycine betaine/carnitine/choline abc transporter;
PDBTitle: crystal structure of glycine betaine/carnitine/choline abc transporter
|
| 54 | c1yewI_ |
|
not modelled |
14.1 |
40 |
PDB header:oxidoreductase, membrane protein
Chain: I: PDB Molecule:particulate methane monooxygenase, b subunit;
PDBTitle: crystal structure of particulate methane monooxygenase
|
| 55 | c2dnfA_ |
|
not modelled |
14.0 |
13 |
PDB header:protein binding
Chain: A: PDB Molecule:doublecortin domain-containing protein 2;
PDBTitle: solution structure of rsgi ruh-062, a dcx domain from human
|
| 56 | d1hyha2 |
|
not modelled |
14.0 |
13 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 57 | d5csma_ |
|
not modelled |
14.0 |
20 |
Fold:Chorismate mutase II
Superfamily:Chorismate mutase II
Family:Allosteric chorismate mutase |
| 58 | d1rxwa1 |
|
not modelled |
13.9 |
20 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 59 | c2j8lB_ |
|
not modelled |
13.9 |
43 |
PDB header:hydrolase
Chain: B: PDB Molecule:coagulation factor xi;
PDBTitle: fxi apple 4 domain loop-out conformation
|
| 60 | d1i39a_ |
|
not modelled |
13.7 |
56 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 61 | c1i3aA_ |
|
not modelled |
13.7 |
56 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease hii;
PDBTitle: rnase hii from archaeoglobus fulgidus with cobalt hexammine2 chloride
|
| 62 | c3k1qS_ |
|
not modelled |
13.6 |
22 |
PDB header:virus
Chain: S: PDB Molecule:vp7: protector protein (outer shell);
PDBTitle: backbone model of an aquareovirus virion by cryo-electron2 microscopy and bioinformatics
|
| 63 | c2hx6A_ |
|
not modelled |
13.6 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease;
PDBTitle: solution structure analysis of the phage t42 endoribonuclease regb
|
| 64 | c2j8lA_ |
|
not modelled |
13.6 |
43 |
PDB header:hydrolase
Chain: A: PDB Molecule:coagulation factor xi;
PDBTitle: fxi apple 4 domain loop-out conformation
|
| 65 | c2j8jA_ |
|
not modelled |
13.6 |
43 |
PDB header:hydrolase
Chain: A: PDB Molecule:coagulation factor xi;
PDBTitle: solution structure of the a4 domain of blood coagulation2 factor xi
|
| 66 | d1k1ga_ |
|
not modelled |
13.5 |
32 |
Fold:Eukaryotic type KH-domain (KH-domain type I)
Superfamily:Eukaryotic type KH-domain (KH-domain type I)
Family:Eukaryotic type KH-domain (KH-domain type I) |
| 67 | d2ldxa2 |
|
not modelled |
13.5 |
25 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 68 | c2kn6A_ |
|
not modelled |
13.5 |
16 |
PDB header:apoptosis
Chain: A: PDB Molecule:apoptosis-associated speck-like protein containing a card;
PDBTitle: structure of full-length human asc (apoptosis-associated speck-like2 protein containing a card)
|
| 69 | d2j0111 |
|
not modelled |
13.4 |
40 |
Fold:L28p-like
Superfamily:L28p-like
Family:Ribosomal protein L28 |
| 70 | d1ekea_ |
|
not modelled |
13.4 |
67 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 71 | c2h8bB_ |
|
not modelled |
13.4 |
50 |
PDB header:hormone/growth factor
Chain: B: PDB Molecule:insulin-like 3;
PDBTitle: solution structure of insl3
|
| 72 | d2axtc1 |
|
not modelled |
13.3 |
18 |
Fold:Photosystem II antenna protein-like
Superfamily:Photosystem II antenna protein-like
Family:Photosystem II antenna protein-like |
| 73 | c2axtc_ |
|
not modelled |
13.3 |
18 |
PDB header:electron transport
Chain: C: PDB Molecule:photosystem ii cp43 protein;
PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
|
| 74 | c2jrpA_ |
|
not modelled |
13.2 |
44 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative cytoplasmic protein;
PDBTitle: solution nmr structure of yfgj from salmonella typhimurium2 modeled with two zn+2 bound, northeast structural genomics3 consortium target str86
|
| 75 | d2oara1 |
|
not modelled |
13.2 |
14 |
Fold:Gated mechanosensitive channel
Superfamily:Gated mechanosensitive channel
Family:Gated mechanosensitive channel |
| 76 | c1sazA_ |
|
not modelled |
13.2 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:probable butyrate kinase 2;
PDBTitle: membership in the askha superfamily: enzymological2 properties and crystal structure of butyrate kinase 2 from3 thermotoga maritima
|
| 77 | d2csba1 |
|
not modelled |
13.1 |
71 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Topoisomerase V repeat domain |
| 78 | c2rejA_ |
|
not modelled |
13.1 |
8 |
PDB header:choline-binding protein
Chain: A: PDB Molecule:putative glycine betaine abc transporter protein;
PDBTitle: abc-transporter choline binding protein in unliganded semi-2 closed conformation
|
| 79 | c2yguA_ |
|
not modelled |
13.0 |
20 |
PDB header:allergen
Chain: A: PDB Molecule:venom allergen 2;
PDBTitle: crystal structure of fire ant venom allergen, sol i 2
|
| 80 | c2jo1A_ |
|
not modelled |
13.0 |
2 |
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
|
| 81 | d2axti1 |
|
not modelled |
13.0 |
23 |
Fold:Single transmembrane helix
Superfamily:Photosystem II reaction center protein I, PsbI
Family:PsbI-like |
| 82 | c3kioA_ |
|
not modelled |
13.0 |
67 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease h2 subunit a;
PDBTitle: mouse rnase h2 complex
|
| 83 | d1l7la_ |
|
not modelled |
13.0 |
14 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:PA-IL, galactose-binding lectin 1 |
| 84 | d1m98a1 |
|
not modelled |
13.0 |
26 |
Fold:Orange carotenoid protein, N-terminal domain
Superfamily:Orange carotenoid protein, N-terminal domain
Family:Orange carotenoid protein, N-terminal domain |
| 85 | c3g9rF_ |
|
not modelled |
12.9 |
36 |
PDB header:viral protein
Chain: F: PDB Molecule:fusion complex of hiv-1 envelope glycoprotein
PDBTitle: structure of the hiv-1 gp41 membrane-proximal ectodomain2 region in a putative prefusion conformation
|
| 86 | c2bbjB_ |
|
not modelled |
12.9 |
15 |
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
|
| 87 | d1uaxa_ |
|
not modelled |
12.8 |
67 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 88 | d1c9ka_ |
|
not modelled |
12.8 |
55 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 89 | c2j8jB_ |
|
not modelled |
12.8 |
43 |
PDB header:hydrolase
Chain: B: PDB Molecule:coagulation factor xi;
PDBTitle: solution structure of the a4 domain of blood coagulation2 factor xi
|
| 90 | d1uf0a_ |
|
not modelled |
12.7 |
17 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Doublecortin (DC)
Family:Doublecortin (DC) |
| 91 | c2p58C_ |
|
not modelled |
12.7 |
45 |
PDB header:transport protein/chaperone
Chain: C: PDB Molecule:putative type iii secretion protein yscg;
PDBTitle: structure of the yersinia pestis type iii secretion system2 needle protein yscf in complex with its chaperones3 ysce/yscg
|
| 92 | c1rr7A_ |
|
not modelled |
12.6 |
29 |
PDB header:transcription
Chain: A: PDB Molecule:middle operon regulator;
PDBTitle: crystal structure of the middle operon regulator protein of2 bacteriophage mu
|
| 93 | d1rr7a_ |
|
not modelled |
12.6 |
29 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Middle operon regulator, Mor |
| 94 | d1j5pa4 |
|
not modelled |
12.6 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 95 | c3s0xB_ |
|
not modelled |
12.6 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptidase a24b, flak domain protein;
PDBTitle: the crystal structure of gxgd membrane protease flak
|
| 96 | c2d0bA_ |
|
not modelled |
12.6 |
44 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease hiii;
PDBTitle: crystal structure of bst-rnase hiii in complex with mg2+
|
| 97 | c2jpmA_ |
|
not modelled |
12.6 |
40 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:bacteriocin lactococcin-g subunit beta;
PDBTitle: lactococcin g-b in tfe
|
| 98 | d1io2a_ |
|
not modelled |
12.5 |
67 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 99 | c2oarA_ |
|
not modelled |
12.4 |
13 |
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
|