| 1 | c1aorB_ |
|
 |
100.0 |
30 |
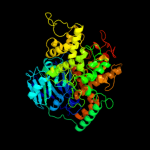
PDB header:oxidoreductase
Chain: B: PDB Molecule:aldehyde ferredoxin oxidoreductase;
PDBTitle: structure of a hyperthermophilic tungstopterin enzyme,2 aldehyde ferredoxin oxidoreductase
|
| 2 | c1b4nD_ |
|
 |
100.0 |
29 |
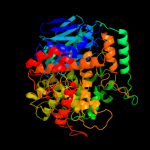
PDB header:oxidoreductase
Chain: D: PDB Molecule:formaldehyde ferredoxin oxidoreductase;
PDBTitle: formaldehyde ferredoxin oxidoreductase from pyrococcus furiosus,2 complexed with glutarate
|
| 3 | d1aora1 |
|
 |
100.0 |
23 |
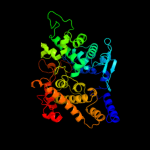
Fold:Aldehyde ferredoxin oxidoreductase, C-terminal domains
Superfamily:Aldehyde ferredoxin oxidoreductase, C-terminal domains
Family:Aldehyde ferredoxin oxidoreductase, C-terminal domains |
| 4 | d1b25a1 |
|
 |
100.0 |
23 |
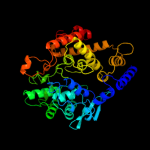
Fold:Aldehyde ferredoxin oxidoreductase, C-terminal domains
Superfamily:Aldehyde ferredoxin oxidoreductase, C-terminal domains
Family:Aldehyde ferredoxin oxidoreductase, C-terminal domains |
| 5 | d1b25a2 |
|
 |
100.0 |
40 |
Fold:Aldehyde ferredoxin oxidoreductase, N-terminal domain
Superfamily:Aldehyde ferredoxin oxidoreductase, N-terminal domain
Family:Aldehyde ferredoxin oxidoreductase, N-terminal domain |
| 6 | d1aora2 |
|
 |
100.0 |
44 |
Fold:Aldehyde ferredoxin oxidoreductase, N-terminal domain
Superfamily:Aldehyde ferredoxin oxidoreductase, N-terminal domain
Family:Aldehyde ferredoxin oxidoreductase, N-terminal domain |
| 7 | d2fug32 |
|
 |
55.6 |
15 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
| 8 | c2e7zA_ |
|
 |
51.8 |
30 |
PDB header:lyase
Chain: A: PDB Molecule:acetylene hydratase ahy;
PDBTitle: acetylene hydratase from pelobacter acetylenicus
|
| 9 | c2vpyE_ |
|
 |
48.4 |
15 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:thiosulfate reductase;
PDBTitle: polysulfide reductase with bound quinone inhibitor,2 pentachlorophenol (pcp)
|
| 10 | c2nyaF_ |
|
 |
47.6 |
15 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:periplasmic nitrate reductase;
PDBTitle: crystal structure of the periplasmic nitrate reductase2 (nap) from escherichia coli
|
| 11 | d1ogya2 |
|
 |
45.9 |
12 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
| 12 | c1ogyA_ |
|
 |
39.2 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:periplasmic nitrate reductase;
PDBTitle: crystal structure of the heterodimeric nitrate reductase2 from rhodobacter sphaeroides
|
| 13 | d1g8ka2 |
|
 |
32.3 |
20 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
| 14 | c1g8jC_ |
|
 |
32.1 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:arsenite oxidase;
PDBTitle: crystal structure analysis of arsenite oxidase from2 alcaligenes faecalis
|
| 15 | c2iv2X_ |
|
 |
30.2 |
22 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:formate dehydrogenase h;
PDBTitle: reinterpretation of reduced form of formate dehydrogenase h2 from e. coli
|
| 16 | d2iv2x2 |
|
 |
29.9 |
19 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
| 17 | c3dfuB_ |
|
 |
29.9 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uncharacterized protein from 6-phosphogluconate
PDBTitle: crystal structure of a putative rossmann-like dehydrogenase (cgl2689)2 from corynebacterium glutamicum at 2.07 a resolution
|
| 18 | d2jioa2 |
|
 |
29.7 |
22 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
| 19 | c2v45A_ |
|
 |
29.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:periplasmic nitrate reductase;
PDBTitle: a new catalytic mechanism of periplasmic nitrate reductase2 from desulfovibrio desulfuricans atcc 27774 from3 crystallographic and epr data and based on detailed4 analysis of the sixth ligand
|
| 20 | c1du9A_ |
|
 |
28.7 |
50 |
PDB header:toxin
Chain: A: PDB Molecule:bmp02 neurotoxin;
PDBTitle: solution structure of bmp02, a natural scorpion toxin which2 blocks apamin-sensitive calcium-activated potassium3 channels, 25 structures
|
| 21 | c1h0hA_ |
|
not modelled |
28.5 |
15 |
PDB header:dehydrogenase
Chain: A: PDB Molecule:formate dehydrogenase (large subunit);
PDBTitle: tungsten containing formate dehydrogenase from2 desulfovibrio gigas
|
| 22 | c2ktcA_ |
|
not modelled |
25.6 |
38 |
PDB header:toxin
Chain: A: PDB Molecule:potassium channel toxin alpha-ktx 9.4;
PDBTitle: solution structure of a novel hkv1.1 inhibiting scorpion toxin from2 mesibuthus tamulus
|
| 23 | c2g5cD_ |
|
not modelled |
25.5 |
22 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from aquifex aeolicus
|
| 24 | c2h2wA_ |
|
not modelled |
24.8 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:homoserine o-succinyltransferase;
PDBTitle: crystal structure of homoserine o-succinyltransferase (ec 2.3.1.46)2 (homoserine o-transsuccinylase) (hts) (tm0881) from thermotoga3 maritima at 2.52 a resolution
|
| 25 | d1yj5a2 |
|
not modelled |
24.3 |
23 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 26 | d2ghra1 |
|
not modelled |
23.4 |
29 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:HTS-like |
| 27 | c3lx4B_ |
|
not modelled |
23.4 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:fe-hydrogenase;
PDBTitle: stepwise [fefe]-hydrogenase h-cluster assembly revealed in the2 structure of hyda(deltaefg)
|
| 28 | c2pv7B_ |
|
not modelled |
22.7 |
31 |
PDB header:isomerase, oxidoreductase
Chain: B: PDB Molecule:t-protein [includes: chorismate mutase (ec 5.4.99.5) (cm)
PDBTitle: crystal structure of chorismate mutase / prephenate dehydrogenase2 (tyra) (1574749) from haemophilus influenzae rd at 2.00 a resolution
|
| 29 | d1u5ta1 |
|
not modelled |
22.0 |
5 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Vacuolar sorting protein domain |
| 30 | c3gmbB_ |
|
not modelled |
20.2 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-methyl-3-hydroxypyridine-5-carboxylic acid
PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic2 acid oxygenase
|
| 31 | c3iufA_ |
|
not modelled |
20.0 |
27 |
PDB header:protein binding
Chain: A: PDB Molecule:zinc finger protein ubi-d4;
PDBTitle: crystal structure of the c2h2-type zinc finger domain of2 human ubi-d4
|
| 32 | c2f1kD_ |
|
not modelled |
19.5 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of synechocystis arogenate dehydrogenase
|
| 33 | c3cgxA_ |
|
not modelled |
19.2 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:putative nucleotide-diphospho-sugar transferase;
PDBTitle: crystal structure of putative nucleotide-diphospho-sugar transferase2 (yp_389115.1) from desulfovibrio desulfuricans g20 at 1.90 a3 resolution
|
| 34 | c2l6aA_ |
|
not modelled |
18.5 |
20 |
PDB header:signaling protein
Chain: A: PDB Molecule:nacht, lrr and pyd domains-containing protein 12;
PDBTitle: three-dimensional structure of the n-terminal effector pyrin domain of2 nlrp12
|
| 35 | d2pv7a2 |
|
not modelled |
18.4 |
31 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 36 | c2vq3B_ |
|
not modelled |
18.2 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:metalloreductase steap3;
PDBTitle: crystal structure of the membrane proximal oxidoreductase2 domain of human steap3, the dominant ferric reductase of3 the erythroid transferrin cycle
|
| 37 | c2fugC_ |
|
not modelled |
18.1 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh-quinone oxidoreductase chain 3;
PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
|
| 38 | c3d1lB_ |
|
not modelled |
17.7 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nadp oxidoreductase bf3122;
PDBTitle: crystal structure of putative nadp oxidoreductase bf3122 from2 bacteroides fragilis
|
| 39 | c1np3B_ |
|
not modelled |
17.1 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: crystal structure of class i acetohydroxy acid isomeroreductase from2 pseudomonas aeruginosa
|
| 40 | d2psba1 |
|
not modelled |
17.0 |
22 |
Fold:YerB-like
Superfamily:YerB-like
Family:YerB-like |
| 41 | c2psbA_ |
|
not modelled |
17.0 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:yerb protein;
PDBTitle: crystal structure of yerb protein from bacillus subtilis.2 northeast structural genomics target sr586
|
| 42 | d1h0ha2 |
|
not modelled |
16.6 |
17 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
| 43 | d2ahra2 |
|
not modelled |
16.1 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 44 | d2f1ka2 |
|
not modelled |
15.8 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 45 | d2pgda2 |
|
not modelled |
15.7 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 46 | d1yqga2 |
|
not modelled |
15.2 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 47 | d1g99a2 |
|
not modelled |
14.8 |
24 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Acetokinase-like |
| 48 | c3dzbA_ |
|
not modelled |
14.7 |
19 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 thermophilus
|
| 49 | c2rafC_ |
|
not modelled |
14.6 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative dinucleotide-binding oxidoreductase;
PDBTitle: crystal structure of putative dinucleotide-binding2 oxidoreductase (np_786167.1) from lactobacillus plantarum3 at 1.60 a resolution
|
| 50 | d2g5ca2 |
|
not modelled |
14.2 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 51 | d2oiea1 |
|
not modelled |
14.1 |
18 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 52 | c3qhaB_ |
|
not modelled |
14.1 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from mycobacterium2 avium 104
|
| 53 | d1f0ya2 |
|
not modelled |
13.3 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 54 | c1z82A_ |
|
not modelled |
13.1 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glycerol-3-phosphate dehydrogenase (tm0378) from2 thermotoga maritima at 2.00 a resolution
|
| 55 | c3ktdC_ |
|
not modelled |
13.0 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of a putative prephenate dehydrogenase (cgl0226)2 from corynebacterium glutamicum atcc 13032 at 2.60 a resolution
|
| 56 | d1kqfa2 |
|
not modelled |
12.9 |
22 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
| 57 | d1vpda2 |
|
not modelled |
12.2 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 58 | c1wxpA_ |
|
not modelled |
12.2 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:tho complex subunit 1;
PDBTitle: solution structure of the death domain of nuclear matrix2 protein p84
|
| 59 | c3o0yC_ |
|
not modelled |
12.1 |
6 |
PDB header:lipid binding protein
Chain: C: PDB Molecule:lipoprotein;
PDBTitle: the crystal structure of the putative lipoprotein from colwellia2 psychrerythraea
|
| 60 | c2ofpB_ |
|
not modelled |
12.1 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketopantoate reductase;
PDBTitle: crystal structure of escherichia coli ketopantoate2 reductase in a ternary complex with nadp+ and pantoate
|
| 61 | d1i36a2 |
|
not modelled |
12.0 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 62 | c3b1fA_ |
|
not modelled |
12.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 mutans
|
| 63 | d1a9xa1 |
|
not modelled |
11.9 |
15 |
Fold:Carbamoyl phosphate synthetase, large subunit connection domain
Superfamily:Carbamoyl phosphate synthetase, large subunit connection domain
Family:Carbamoyl phosphate synthetase, large subunit connection domain |
| 64 | c1ks9A_ |
|
not modelled |
11.7 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: ketopantoate reductase from escherichia coli
|
| 65 | c2axoA_ |
|
not modelled |
11.3 |
83 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein atu2684;
PDBTitle: x-ray crystal structure of protein agr_c_4864 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr35.
|
| 66 | c1i36A_ |
|
not modelled |
11.3 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein mth1747;
PDBTitle: structure of conserved protein mth1747 of unknown function2 reveals structural similarity with 3-hydroxyacid3 dehydrogenases
|
| 67 | d2axoa1 |
|
not modelled |
11.2 |
83 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Atu2684-like |
| 68 | c2fpgA_ |
|
not modelled |
10.9 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa ligase [gdp-forming] alpha-chain,
PDBTitle: crystal structure of pig gtp-specific succinyl-coa2 synthetase in complex with gdp
|
| 69 | c1wcnA_ |
|
not modelled |
10.9 |
35 |
PDB header:rna-binding protein
Chain: A: PDB Molecule:transcription elongation protein nusa;
PDBTitle: nmr structure of the carboxyterminal domains of escherichia2 coli nusa
|
| 70 | c2uyyD_ |
|
not modelled |
10.9 |
15 |
PDB header:cytokine
Chain: D: PDB Molecule:n-pac protein;
PDBTitle: structure of the cytokine-like nuclear factor n-pac
|
| 71 | c3k3qB_ |
|
not modelled |
10.8 |
13 |
PDB header:immune system
Chain: B: PDB Molecule:botulinum neurotoxin type a;
PDBTitle: crystal structure of a llama antibody complexed with the c.2 botulinum neurotoxin serotype a catalytic domain
|
| 72 | c3g17H_ |
|
not modelled |
10.5 |
12 |
PDB header:structural genomics, unknown function
Chain: H: PDB Molecule:similar to 2-dehydropantoate 2-reductase;
PDBTitle: structure of putative 2-dehydropantoate 2-reductase from2 staphylococcus aureus
|
| 73 | d2jeka1 |
|
not modelled |
10.3 |
32 |
Fold:Rv1873-like
Superfamily:Rv1873-like
Family:Rv1873-like |
| 74 | c2howB_ |
|
not modelled |
10.3 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:356aa long hypothetical dipeptidase;
PDBTitle: dipeptidase (ph0974) from pyrococcus horikoshii ot3
|
| 75 | c3idwA_ |
|
not modelled |
10.3 |
39 |
PDB header:endocytosis
Chain: A: PDB Molecule:actin cytoskeleton-regulatory complex protein sla1;
PDBTitle: crystal structure of sla1 homology domain 2
|
| 76 | c3qf2B_ |
|
not modelled |
10.1 |
15 |
PDB header:apoptosis
Chain: B: PDB Molecule:nacht, lrr and pyd domains-containing protein 3;
PDBTitle: crystal structure of nalp3 pyd
|
| 77 | d1bg6a2 |
|
not modelled |
10.0 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 78 | c1kqgA_ |
|
not modelled |
9.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:formate dehydrogenase, nitrate-inducible, major subunit;
PDBTitle: formate dehydrogenase n from e. coli
|
| 79 | c2eapA_ |
|
not modelled |
9.7 |
19 |
PDB header:signaling protein
Chain: A: PDB Molecule:lymphocyte cytosolic protein 2;
PDBTitle: solution structure of the n-terminal sam-domain of human2 lymphocyte cytosolic protein 2
|
| 80 | c2yx6C_ |
|
not modelled |
9.6 |
13 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hypothetical protein ph0822;
PDBTitle: crystal structure of ph0822
|
| 81 | d1af7a1 |
|
not modelled |
9.5 |
16 |
Fold:Chemotaxis receptor methyltransferase CheR, N-terminal domain
Superfamily:Chemotaxis receptor methyltransferase CheR, N-terminal domain
Family:Chemotaxis receptor methyltransferase CheR, N-terminal domain |
| 82 | c1e1hC_ |
|
not modelled |
9.4 |
13 |
PDB header:hydrolase
Chain: C: PDB Molecule:botulinum neurotoxin type a light chain;
PDBTitle: crystal structure of recombinant botulinum neurotoxin type2 a light chain, self-inhibiting zn endopeptidase.
|
| 83 | c3ju3A_ |
|
not modelled |
9.3 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 2-oxoacid ferredoxin oxidoreductase, alpha chain;
PDBTitle: crystal structure of alpha chain of probable 2-oxoacid ferredoxin2 oxidoreductase from thermoplasma acidophilum
|
| 84 | c3i83B_ |
|
not modelled |
9.3 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of 2-dehydropantoate 2-reductase from methylococcus2 capsulatus
|
| 85 | d1wdka3 |
|
not modelled |
9.3 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 86 | c3bd1B_ |
|
not modelled |
9.2 |
14 |
PDB header:transcription
Chain: B: PDB Molecule:cro protein;
PDBTitle: structure of the cro protein from putative prophage element xfaso 1 in2 xylella fastidiosa strain ann-1
|
| 87 | c2q4pA_ |
|
not modelled |
9.0 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein rs21-c6;
PDBTitle: ensemble refinement of the crystal structure of protein from mus2 musculus mm.29898
|
| 88 | d2a3qa1 |
|
not modelled |
9.0 |
18 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 89 | c2kn6A_ |
|
not modelled |
9.0 |
28 |
PDB header:apoptosis
Chain: A: PDB Molecule:apoptosis-associated speck-like protein containing a card;
PDBTitle: structure of full-length human asc (apoptosis-associated speck-like2 protein containing a card)
|
| 90 | d1trba1 |
|
not modelled |
9.0 |
15 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 91 | c3k96B_ |
|
not modelled |
8.9 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
|
| 92 | c3ewiB_ |
|
not modelled |
8.9 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:n-acylneuraminate cytidylyltransferase;
PDBTitle: structural analysis of the c-terminal domain of murine cmp-2 sialic acid synthetase
|
| 93 | c3iukB_ |
|
not modelled |
8.9 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of putative bacterial protein of unknown function2 (duf885, pf05960.1, ) from arthrobacter aurescens tc1, reveals fold3 similar to that of m32 carboxypeptidases
|
| 94 | d3c8ya1 |
|
not modelled |
8.8 |
14 |
Fold:Fe-only hydrogenase
Superfamily:Fe-only hydrogenase
Family:Fe-only hydrogenase |
| 95 | c2vouA_ |
|
not modelled |
8.8 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,6-dihydroxypyridine hydroxylase;
PDBTitle: structure of 2,6-dihydroxypyridine-3-hydroxylase from2 arthrobacter nicotinovorans
|
| 96 | c3h0mE_ |
|
not modelled |
8.7 |
15 |
PDB header:ligase
Chain: E: PDB Molecule:aspartyl/glutamyl-trna(asn/gln) amidotransferase
PDBTitle: structure of trna-dependent amidotransferase gatcab from2 aquifex aeolicus
|
| 97 | d1nvma1 |
|
not modelled |
8.6 |
33 |
Fold:RuvA C-terminal domain-like
Superfamily:post-HMGL domain-like
Family:DmpG/LeuA communication domain-like |
| 98 | d1y5ia2 |
|
not modelled |
8.6 |
10 |
Fold:Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily:Formate dehydrogenase/DMSO reductase, domains 1-3
Family:Formate dehydrogenase/DMSO reductase, domains 1-3 |
| 99 | c2p2vA_ |
|
not modelled |
8.5 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:alpha-2,3-sialyltransferase;
PDBTitle: crystal structure analysis of monofunctional alpha-2,3-2 sialyltransferase cst-i from campylobacter jejuni
|

