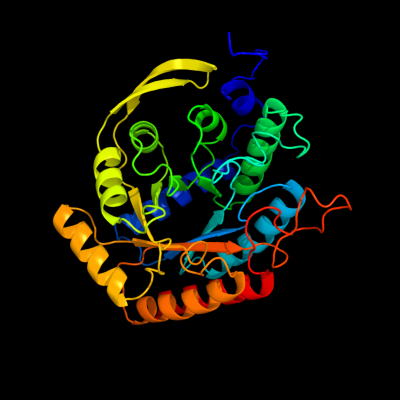
| 1 | d1n8fa_
|
|
 |
100.0 |
57 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
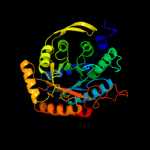
| 2 | c3tqkA_
|
|
 |
100.0 |
46 |
PDB header:transferase
Chain: A: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: structure of phospho-2-dehydro-3-deoxyheptonate aldolase from2 francisella tularensis schu s4
|
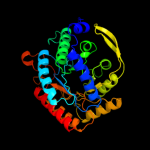
| 3 | d1of8a_
|
|
 |
100.0 |
51 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
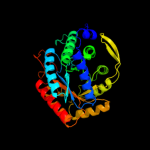
| 4 | c1ofaB_
|
|
 |
100.0 |
51 |
PDB header:lyase
Chain: B: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: crystal structure of the tyrosine-regulated2 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase3 from saccharomyces cerevisiae in complex with4 phosphoenolpyruvate and cobalt(ii)
|
| 5 | d1vr6a1
|
|
 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 6 | c1vs1B_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:3-deoxy-7-phosphoheptulonate synthase;
PDBTitle: crystal structure of 3-deoxy-d-arabino-heptulosonate-7-2 phosphate synthase (dahp synthase) from aeropyrum pernix3 in complex with mn2+ and pep
|
| 7 | c1zcoA_
|
|
 |
100.0 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphoheptonate aldolase;
PDBTitle: crystal structure of pyrococcus furiosus 3-deoxy-d-arabino-2 heptulosonate 7-phosphate synthase
|
| 8 | d2a21a1
|
|
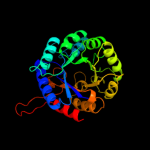 |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 9 | c3nvtA_
|
|
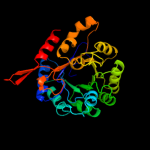 |
100.0 |
19 |
PDB header:transferase/isomerase
Chain: A: PDB Molecule:3-deoxy-d-arabino-heptulosonate 7-phosphate synthase;
PDBTitle: 1.95 angstrom crystal structure of a bifunctional 3-deoxy-7-2 phosphoheptulonate synthase/chorismate mutase (aroa) from listeria3 monocytogenes egd-e
|
| 10 | c3stgA_
|
|
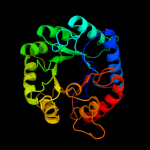 |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of a58p, del(n59), and loop 7 truncated mutant of 3-2 deoxy-d-manno-octulosonate 8-phosphate synthase (kdo8ps) from3 neisseria meningitidis
|
| 11 | c3fs2A_
|
|
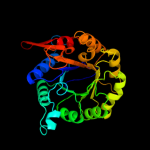 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate2 aldolase from bruciella melitensis at 1.85a resolution
|
| 12 | d1d9ea_
|
|
 |
100.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 13 | d1o60a_
|
|
 |
100.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 14 | c3sz8D_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 2;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
|
| 15 | c3t4cD_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 1;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia ambifaria
|
| 16 | c3pg8B_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: truncated form of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase2 from thermotoga maritima
|
| 17 | d2zdra2
|
|
 |
99.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 18 | d1vlia2
|
|
 |
99.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 19 | c1xuzA_
|
|
 |
98.7 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:polysialic acid capsule biosynthesis protein siac;
PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
|
| 20 | d2b7oa1
|
|
 |
98.1 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class-II DAHP synthetase |
| 21 | c1vliA_ |
|
not modelled |
98.0 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:spore coat polysaccharide biosynthesis protein spse;
PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
|
| 22 | c3g8rA_ |
|
not modelled |
97.5 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:probable spore coat polysaccharide biosynthesis protein e;
PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
|
| 23 | d1ajza_ |
|
not modelled |
93.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 24 | d1ad1a_ |
|
not modelled |
93.5 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 25 | c2y5sA_ |
|
not modelled |
86.6 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
|
| 26 | c3eb2A_ |
|
not modelled |
84.2 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 27 | d1dxha1 |
|
not modelled |
82.5 |
22 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 28 | c2otcA_ |
|
not modelled |
79.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: ornithine transcarbamoylase complexed with n-2 (phosphonacetyl)-l-ornithine
|
| 29 | c3tr9A_ |
|
not modelled |
79.2 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
| 30 | c1ortD_ |
|
not modelled |
76.3 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine transcarbamoylase;
PDBTitle: ornithine transcarbamoylase from pseudomonas aeruginosa
|
| 31 | c1tx2A_ |
|
not modelled |
74.6 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
| 32 | d1tx2a_ |
|
not modelled |
74.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 33 | c1ml4A_ |
|
not modelled |
71.2 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:aspartate transcarbamoylase;
PDBTitle: the pala-liganded aspartate transcarbamoylase catalytic subunit from2 pyrococcus abyssi
|
| 34 | d1sq5a_ |
|
not modelled |
70.8 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Phosphoribulokinase/pantothenate kinase |
| 35 | c3updA_ |
|
not modelled |
69.7 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: 2.9 angstrom crystal structure of ornithine carbamoyltransferase2 (argf) from vibrio vulnificus
|
| 36 | d1duvg1 |
|
not modelled |
69.0 |
22 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 37 | c2vqqA_ |
|
not modelled |
64.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:histone deacetylase 4;
PDBTitle: structure of hdac4 catalytic domain (a double cysteine-to-2 alanine mutant) bound to a trifluoromethylketone inhbitor
|
| 38 | d1eyea_ |
|
not modelled |
64.6 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 39 | c3d6nB_ |
|
not modelled |
63.8 |
20 |
PDB header:hydrolase/transferase
Chain: B: PDB Molecule:aspartate carbamoyltransferase;
PDBTitle: crystal structure of aquifex dihydroorotase activated by aspartate2 transcarbamoylase
|
| 40 | c3tqcB_ |
|
not modelled |
60.0 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:pantothenate kinase;
PDBTitle: structure of the pantothenate kinase (coaa) from coxiella burnetii
|
| 41 | c3e5bB_ |
|
not modelled |
59.6 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:isocitrate lyase;
PDBTitle: 2.4 a crystal structure of isocitrate lyase from brucella2 melitensis
|
| 42 | d1vlva1 |
|
not modelled |
57.9 |
16 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 43 | d1f74a_ |
|
not modelled |
57.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 44 | d1ml4a1 |
|
not modelled |
56.8 |
24 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 45 | d1otha1 |
|
not modelled |
56.3 |
22 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 46 | c2p2gD_ |
|
not modelled |
56.2 |
28 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from mycobacterium2 tuberculosis (rv1656): orthorhombic form
|
| 47 | c2e3zB_ |
|
not modelled |
56.1 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of intracellular family 1 beta-2 glucosidase bgl1a from the basidiomycete phanerochaete3 chrysosporium in substrate-free form
|
| 48 | c3gd5D_ |
|
not modelled |
54.3 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from gloeobacter2 violaceus
|
| 49 | c1a1sA_ |
|
not modelled |
52.5 |
21 |
PDB header:transcarbamylase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: ornithine carbamoyltransferase from pyrococcus furiosus
|
| 50 | c3si9B_ |
|
not modelled |
50.9 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
|
| 51 | d1o5ka_ |
|
not modelled |
49.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 52 | d1pvva1 |
|
not modelled |
49.7 |
20 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 53 | c3grfA_ |
|
not modelled |
47.8 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: x-ray structure of ornithine transcarbamoylase from giardia2 lamblia
|
| 54 | c1vlvA_ |
|
not modelled |
47.2 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase (tm1097) from2 thermotoga maritima at 2.25 a resolution
|
| 55 | c2ze5A_ |
|
not modelled |
47.1 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:isopentenyl transferase;
PDBTitle: crystal structure of adenosine phosphate-isopentenyltransferase
|
| 56 | c3bi8A_ |
|
not modelled |
47.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
|
| 57 | c3ptkB_ |
|
not modelled |
46.2 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase os4bglu12;
PDBTitle: the crystal structure of rice (oryza sativa l.) os4bglu12
|
| 58 | c2w37A_ |
|
not modelled |
45.5 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase, catabolic;
PDBTitle: crystal structure of the hexameric catabolic ornithine2 transcarbamylase from lactobacillus hilgardii
|
| 59 | c3pueA_ |
|
not modelled |
44.8 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
| 60 | c1fvoB_ |
|
not modelled |
44.5 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:ornithine transcarbamylase;
PDBTitle: crystal structure of human ornithine transcarbamylase complexed with2 carbamoyl phosphate
|
| 61 | d1tuga1 |
|
not modelled |
43.8 |
19 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 62 | c2ef0A_ |
|
not modelled |
41.3 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from thermus2 thermophilus
|
| 63 | c2hjpA_ |
|
not modelled |
40.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
| 64 | d2e1da1 |
|
not modelled |
38.6 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 65 | c2p2sA_ |
|
not modelled |
38.1 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase (yp_050235.1) from2 erwinia carotovora atroseptica scri1043 at 1.25 a resolution
|
| 66 | c3a8tA_ |
|
not modelled |
37.6 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:adenylate isopentenyltransferase;
PDBTitle: plant adenylate isopentenyltransferase in complex with atp
|
| 67 | c3ahyD_ |
|
not modelled |
36.9 |
10 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of beta-glucosidase 2 from fungus trichoderma reesei2 in complex with tris
|
| 68 | c2ehhE_ |
|
not modelled |
36.3 |
19 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
| 69 | c3noeA_ |
|
not modelled |
36.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
| 70 | c3daqB_ |
|
not modelled |
35.3 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
| 71 | c2bmbA_ |
|
not modelled |
35.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:folic acid synthesis protein fol1;
PDBTitle: x-ray structure of the bifunctional 6-hydroxymethyl-7,8-2 dihydroxypterin pyrophosphokinase dihydropteroate synthase3 from saccharomyces cerevisiae
|
| 72 | c3cprB_ |
|
not modelled |
33.6 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthetase;
PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
|
| 73 | d2v3za1 |
|
not modelled |
33.4 |
21 |
Fold:Ribonuclease H-like motif
Superfamily:Creatinase/prolidase N-terminal domain
Family:Creatinase/prolidase N-terminal domain |
| 74 | d1u1ia1 |
|
not modelled |
33.0 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 75 | d1igwa_ |
|
not modelled |
33.0 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 76 | c3b4uB_ |
|
not modelled |
32.0 |
7 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from agrobacterium2 tumefaciens str. c58
|
| 77 | d2v89a1 |
|
not modelled |
31.5 |
21 |
Fold:FYVE/PHD zinc finger
Superfamily:FYVE/PHD zinc finger
Family:PHD domain |
| 78 | c3d3qB_ |
|
not modelled |
29.1 |
28 |
PDB header:transferase
Chain: B: PDB Molecule:trna delta(2)-isopentenylpyrophosphate
PDBTitle: crystal structure of trna delta(2)-isopentenylpyrophosphate2 transferase (se0981) from staphylococcus epidermidis.3 northeast structural genomics consortium target ser100
|
| 79 | d1pffa_ |
|
not modelled |
29.1 |
25 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 80 | c2jf7B_ |
|
not modelled |
28.7 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:strictosidine-o-beta-d-glucosidase;
PDBTitle: structure of strictosidine glucosidase
|
| 81 | c2yxgD_ |
|
not modelled |
28.3 |
19 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
| 82 | d1ulza3 |
|
not modelled |
28.3 |
11 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:BC ATP-binding domain-like |
| 83 | c3u57A_ |
|
not modelled |
28.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:raucaffricine-o-beta-d-glucosidase;
PDBTitle: structures of alkaloid biosynthetic glucosidases decode substrate2 specificity
|
| 84 | c1h6dL_ |
|
not modelled |
27.8 |
18 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
| 85 | c3exaD_ |
|
not modelled |
27.1 |
28 |
PDB header:transferase
Chain: D: PDB Molecule:trna delta(2)-isopentenylpyrophosphate
PDBTitle: crystal structure of the full-length trna2 isopentenylpyrophosphate transferase (bh2366) from3 bacillus halodurans, northeast structural genomics4 consortium target bhr41.
|
| 86 | c2qt7B_ |
|
not modelled |
26.9 |
36 |
PDB header:hydrolase
Chain: B: PDB Molecule:receptor-type tyrosine-protein phosphatase-like
PDBTitle: crystallographic structure of the mature ectodomain of the2 human receptor-type protein-tyrosine phosphatase ia-2 at3 1.30 angstroms
|
| 87 | c1v02F_ |
|
not modelled |
26.8 |
21 |
PDB header:hydrolase
Chain: F: PDB Molecule:dhurrinase;
PDBTitle: crystal structure of the sorghum bicolor dhurrinase 1
|
| 88 | d1v02a_ |
|
not modelled |
26.8 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 89 | c3fluD_ |
|
not modelled |
26.6 |
18 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 90 | c3oqbF_ |
|
not modelled |
26.0 |
20 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from bradyrhizobium2 japonicum usda 110
|
| 91 | c3na8A_ |
|
not modelled |
25.4 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
|
| 92 | c2ixaA_ |
|
not modelled |
24.4 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-n-acetylgalactosaminidase;
PDBTitle: a-zyme, n-acetylgalactosaminidase
|
| 93 | d2auwa1 |
|
not modelled |
24.4 |
75 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:NE0471 C-terminal domain-like |
| 94 | c2h9aA_ |
|
not modelled |
24.3 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:carbon monoxide dehydrogenase corrinoid/iron-
PDBTitle: corrinoid iron-sulfur protein
|
| 95 | c1ofgF_ |
|
not modelled |
24.3 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
| 96 | c3btuD_ |
|
not modelled |
23.0 |
18 |
PDB header:transcription
Chain: D: PDB Molecule:galactose/lactose metabolism regulatory protein
PDBTitle: crystal structure of the super-repressor mutant of gal80p2 from saccharomyces cerevisiae; gal80(s2) [e351k]
|
| 97 | c2wamB_ |
|
not modelled |
22.6 |
13 |
PDB header:unknown function
Chain: B: PDB Molecule:conserved hypothetical alanine and leucine rich
PDBTitle: crystal structure of mycobacterium tuberculosis unknown2 function protein rv2714
|
| 98 | c2auwB_ |
|
not modelled |
22.5 |
75 |
PDB header:unknown function
Chain: B: PDB Molecule:hypothetical protein ne0471;
PDBTitle: crystal structure of putative dna binding protein ne0471 from2 nitrosomonas europaea atcc 19718
|
| 99 | d1ryda1 |
|
not modelled |
21.9 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 100 | c3crqA_ |
|
not modelled |
21.8 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:trna delta(2)-isopentenylpyrophosphate
PDBTitle: structure of trna dimethylallyltransferase: rna2 modification through a channel
|
| 101 | c3cdzB_ |
|
not modelled |
21.7 |
10 |
PDB header:blood clotting
Chain: B: PDB Molecule:coagulation factor viii light chain;
PDBTitle: crystal structure of human factor viii
|
| 102 | c3g0sA_ |
|
not modelled |
21.7 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from salmonella typhimurium lt2
|
| 103 | c3fhlC_ |
|
not modelled |
21.4 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from bacteroides2 fragilis nctc 9343
|
| 104 | c3fa4D_ |
|
not modelled |
21.4 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
| 105 | d1ydwa1 |
|
not modelled |
21.2 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 106 | c3fozB_ |
|
not modelled |
21.1 |
20 |
PDB header:transferase/rna
Chain: B: PDB Molecule:trna delta(2)-isopentenylpyrophosphate transferase;
PDBTitle: structure of e. coli isopentenyl-trna transferase in complex with e.2 coli trna(phe)
|
| 107 | d1f05a_ |
|
not modelled |
20.6 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 108 | c3m16A_ |
|
not modelled |
20.4 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:transaldolase;
PDBTitle: structure of a transaldolase from oleispira antarctica
|