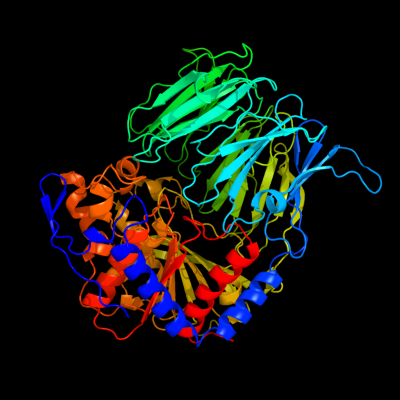
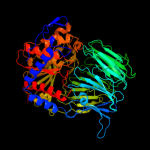
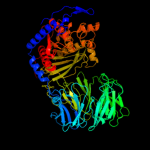
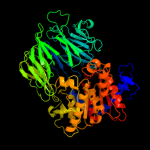
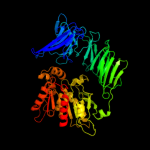
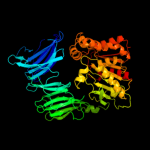
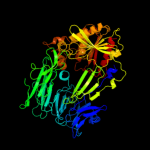
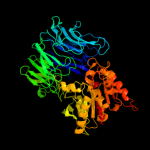
1 c2xe4A_
100.0
37
PDB header: hydrolase/inhibitorChain: A: PDB Molecule: oligopeptidase b;PDBTitle: structure of oligopeptidase b from leishmania major
2 c1yr2A_
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: prolyl oligopeptidase;PDBTitle: structural and mechanistic analysis of two prolyl endopeptidases: role2 of inter-domain dynamics in catalysis and specificity
3 c2bklB_
100.0
26
PDB header: hydrolaseChain: B: PDB Molecule: prolyl endopeptidase;PDBTitle: structural and mechanistic analysis of two prolyl2 endopeptidases: role of inter-domain dynamics in3 catalysis and specificity
4 c3iumA_
100.0
23
PDB header: hydrolaseChain: A: PDB Molecule: prolyl endopeptidase;PDBTitle: appep_wtx opened state
5 c1qfmA_
100.0
24
PDB header: hydrolaseChain: A: PDB Molecule: protein (prolyl oligopeptidase);PDBTitle: prolyl oligopeptidase from porcine muscle
6 c2ecfA_
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: dipeptidyl peptidase iv;PDBTitle: crystal structure of dipeptidyl aminopeptidase iv from2 stenotrophomonas maltophilia
7 c2g5tA_
100.0
14
PDB header: hydrolaseChain: A: PDB Molecule: dipeptidyl peptidase 4;PDBTitle: crystal structure of human dipeptidyl peptidase iv (dppiv)2 complexed with cyanopyrrolidine (c5-pro-pro) inhibitor 21ag
8 c2eepA_
100.0
14
PDB header: hydrolaseChain: A: PDB Molecule: dipeptidyl aminopeptidase iv, putative;PDBTitle: prolyl tripeptidyl aminopeptidase complexed with an inhibitor
9 c2qtbB_
100.0
15
PDB header: hydrolaseChain: B: PDB Molecule: dipeptidyl peptidase 4;PDBTitle: human dipeptidyl peptidase iv/cd26 in complex with a 4-aryl2 cyclohexylalanine inhibitor
10 c1z68A_
100.0
16
PDB header: lyaseChain: A: PDB Molecule: fibroblast activation protein, alpha subunit;PDBTitle: crystal structure of human fibroblast activation protein alpha
11 c3azqA_
100.0
14
PDB header: hydrolaseChain: A: PDB Molecule: aminopeptidase;PDBTitle: crystal structure of puromycin hydrolase s511a mutant complexed with2 pgg
12 c2hu7A_
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: acylamino-acid-releasing enzyme;PDBTitle: binding of inhibitors by acylaminoacyl peptidase
13 c1xfdD_
100.0
13
PDB header: membrane proteinChain: D: PDB Molecule: dipeptidyl aminopeptidase-like protein 6;PDBTitle: structure of a human a-type potassium channel accelerating factor2 dppx, a member of the dipeptidyl aminopeptidase family
14 d1qfma1
100.0
19
Fold: 7-bladed beta-propellerSuperfamily: Peptidase/esterase 'gauge' domainFamily: Prolyl oligopeptidase, N-terminal domain15 d1qfma2
100.0
29
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Prolyl oligopeptidase, C-terminal domain16 c3doiA_
100.0
14
PDB header: hydrolaseChain: A: PDB Molecule: esterase;PDBTitle: crystal structure of a thermostable esterase complex with2 paraoxon
17 d2hu7a2
99.9
20
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acylamino-acid-releasing enzyme, C-terminal donain18 d1orva2
99.9
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: DPP6 catalytic domain-like19 d2bgra2
99.9
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: DPP6 catalytic domain-like20 c3k2iA_
99.9
11
PDB header: hydrolaseChain: A: PDB Molecule: acyl-coenzyme a thioesterase 4;PDBTitle: human acyl-coenzyme a thioesterase 4
21 d2jbwa1
not modelled
99.9
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: 2,6-dihydropseudooxynicotine hydrolase-like22 c2jbwB_
not modelled
99.9
17
PDB header: hydrolaseChain: B: PDB Molecule: 2,6-dihydroxy-pseudo-oxynicotine hydrolase;PDBTitle: crystal structure of the 2,6-dihydroxy-pseudo-oxynicotine2 hydrolase.
23 d1vlqa_
not modelled
99.9
12
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetyl xylan esterase-like24 d2b9va2
not modelled
99.9
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: PepX catalytic domain-like25 c3fnbB_
not modelled
99.9
13
PDB header: hydrolaseChain: B: PDB Molecule: acylaminoacyl peptidase smu_737;PDBTitle: crystal structure of acylaminoacyl peptidase smu_737 from2 streptococcus mutans ua159
26 d1xfda2
not modelled
99.8
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: DPP6 catalytic domain-like27 c3hlkB_
not modelled
99.8
13
PDB header: hydrolaseChain: B: PDB Molecule: acyl-coenzyme a thioesterase 2, mitochondrial;PDBTitle: crystal structure of human mitochondrial acyl-coa2 thioesterase (acot2)
28 d1mpxa2
not modelled
99.8
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: PepX catalytic domain-like29 c2b9vB_
not modelled
99.8
14
PDB header: hydrolaseChain: B: PDB Molecule: alpha-amino acid ester hydrolase;PDBTitle: acetobacter turbidans alpha-amino acid ester hydrolase
30 d1ju3a2
not modelled
99.8
21
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: PepX catalytic domain-like31 c1l7qA_
not modelled
99.8
21
PDB header: hydrolaseChain: A: PDB Molecule: cocaine esterase;PDBTitle: ser117ala mutant of bacterial cocaine esterase coce
32 c3ls2D_
not modelled
99.8
14
PDB header: hydrolaseChain: D: PDB Molecule: s-formylglutathione hydrolase;PDBTitle: crystal structure of an s-formylglutathione hydrolase from2 pseudoalteromonas haloplanktis tac125
33 d1l7aa_
not modelled
99.8
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetyl xylan esterase-like34 c3h2iA_
not modelled
99.8
15
PDB header: hydrolaseChain: A: PDB Molecule: esterase;PDBTitle: crystal structure of n228w mutant of the rice cell wall2 degrading esterase lipa from xanthomonas oryzae
35 c3bxpA_
not modelled
99.8
9
PDB header: hydrolaseChain: A: PDB Molecule: putative lipase/esterase;PDBTitle: crystal structure of a putative carboxylesterase (lp_2923) from2 lactobacillus plantarum wcfs1 at 1.70 a resolution
36 c2o2gA_
not modelled
99.8
17
PDB header: hydrolaseChain: A: PDB Molecule: dienelactone hydrolase;PDBTitle: crystal structure of dienelactone hydrolase (yp_324580.1) from2 anabaena variabilis atcc 29413 at 1.92 a resolution
37 d1lnsa3
not modelled
99.8
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: PepX catalytic domain-like38 c2veoA_
not modelled
99.8
17
PDB header: hydrolaseChain: A: PDB Molecule: lipase a;PDBTitle: x-ray structure of candida antarctica lipase a in its2 closed state.
39 c3ib3A_
not modelled
99.8
14
PDB header: hydrolaseChain: A: PDB Molecule: coce/nond family hydrolase;PDBTitle: crystal structure of sacol2612 - coce/nond family hydrolase from2 staphylococcus aureus
40 c3i6yA_
not modelled
99.8
13
PDB header: hydrolaseChain: A: PDB Molecule: esterase apc40077;PDBTitle: structure of an esterase from the oil-degrading bacterium oleispira2 antarctica
41 c1mpxB_
not modelled
99.8
17
PDB header: hydrolaseChain: B: PDB Molecule: alpha-amino acid ester hydrolase;PDBTitle: alpha-amino acid ester hydrolase labeled with selenomethionine
42 d1jkma_
not modelled
99.8
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase43 c3fcxA_
not modelled
99.8
15
PDB header: hydrolaseChain: A: PDB Molecule: s-formylglutathione hydrolase;PDBTitle: crystal structure of human esterase d
44 c3mveB_
not modelled
99.8
11
PDB header: lyaseChain: B: PDB Molecule: upf0255 protein vv1_0328;PDBTitle: crystal structure of a novel pyruvate decarboxylase
45 c3ksrA_
not modelled
99.8
14
PDB header: hydrolaseChain: A: PDB Molecule: putative serine hydrolase;PDBTitle: crystal structure of a putative serine hydrolase (xcc3885) from2 xanthomonas campestris pv. campestris at 2.69 a resolution
46 c2zshA_
not modelled
99.7
15
PDB header: hormone receptorChain: A: PDB Molecule: probable gibberellin receptor gid1l1;PDBTitle: structural basis of gibberellin(ga3)-induced della2 recognition by the gibberellin receptor
47 c3ed1E_
not modelled
99.7
12
PDB header: hydrolase receptorChain: E: PDB Molecule: gibberellin receptor gid1;PDBTitle: crystal structure of rice gid1 complexed with ga3
48 c2o7vA_
not modelled
99.7
17
PDB header: hydrolaseChain: A: PDB Molecule: cxe carboxylesterase;PDBTitle: carboxylesterase aecxe1 from actinidia eriantha covalently inhibited2 by paraoxon
49 c3f67A_
not modelled
99.7
13
PDB header: hydrolaseChain: A: PDB Molecule: putative dienelactone hydrolase;PDBTitle: crystal structure of putative dienelactone hydrolase from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
50 c3fcyB_
not modelled
99.7
15
PDB header: hydrolaseChain: B: PDB Molecule: xylan esterase 1;PDBTitle: crystal structure of acetyl xylan esterase 1 from2 thermoanaerobacterium sp. jw/sl ys485
51 d1sfra_
not modelled
99.7
13
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Mycobacterial antigens52 c2i3dA_
not modelled
99.7
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein atu1826;PDBTitle: crystal structure of protein of unknown function atu1826, a putative2 alpha/beta hydrolase from agrobacterium tumefaciens
53 d2i3da1
not modelled
99.7
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Atu1826-like54 c3e4dD_
not modelled
99.7
14
PDB header: hydrolaseChain: D: PDB Molecule: esterase d;PDBTitle: structural and kinetic study of an s-formylglutathione2 hydrolase from agrobacterium tumefaciens
55 c3qh4A_
not modelled
99.7
18
PDB header: hydrolaseChain: A: PDB Molecule: esterase lipw;PDBTitle: crystal structure of esterase lipw from mycobacterium marinum
56 c3d0kA_
not modelled
99.7
13
PDB header: hydrolaseChain: A: PDB Molecule: putative poly(3-hydroxybutyrate) depolymerase lpqc;PDBTitle: crystal structure of the lpqc, poly(3-hydroxybutyrate) depolymerase2 from bordetella parapertussis
57 c3ga7A_
not modelled
99.7
10
PDB header: hydrolaseChain: A: PDB Molecule: acetyl esterase;PDBTitle: 1.55 angstrom crystal structure of an acetyl esterase from salmonella2 typhimurium
58 c2wtmC_
not modelled
99.7
18
PDB header: hydrolaseChain: C: PDB Molecule: est1e;PDBTitle: est1e from butyrivibrio proteoclasticus
59 d1pv1a_
not modelled
99.7
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hypothetical esterase YJL068C60 d1lzla_
not modelled
99.7
20
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase61 d1jjfa_
not modelled
99.7
12
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase62 d1dina_
not modelled
99.7
20
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Dienelactone hydrolase63 d1f0na_
not modelled
99.7
10
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Mycobacterial antigens64 d1dqza_
not modelled
99.7
12
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Mycobacterial antigens65 d2fuka1
not modelled
99.7
18
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Atu1826-like66 c1lnsA_
not modelled
99.7
17
PDB header: hydrolaseChain: A: PDB Molecule: x-prolyl dipeptidyl aminopetidase;PDBTitle: crystal structure analysis of the x-prolyl dipeptidyl2 aminopeptidase from lactococcus lactis
67 d2gzsa1
not modelled
99.7
13
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: IroE-like68 c3trdA_
not modelled
99.6
15
PDB header: hydrolaseChain: A: PDB Molecule: alpha/beta hydrolase;PDBTitle: structure of an alpha-beta serine hydrolase homologue from coxiella2 burnetii
69 c3hxkB_
not modelled
99.6
16
PDB header: hydrolaseChain: B: PDB Molecule: sugar hydrolase;PDBTitle: crystal structure of a sugar hydrolase (yeeb) from2 lactococcus lactis, northeast structural genomics3 consortium target kr108
70 c3d7rB_
not modelled
99.6
13
PDB header: hydrolaseChain: B: PDB Molecule: esterase;PDBTitle: crystal structure of a putative esterase from staphylococcus aureus
71 c3dnmA_
not modelled
99.6
15
PDB header: hydrolaseChain: A: PDB Molecule: esterase/lipase;PDBTitle: crystal structure hormone-sensitive lipase from a2 metagenome library
72 c2qm0B_
not modelled
99.6
10
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: bes;PDBTitle: crystal structure of bes protein from bacillus cereus
73 c3fakA_
not modelled
99.6
15
PDB header: hydrolaseChain: A: PDB Molecule: esterase/lipase;PDBTitle: structural and functional analysis of a hormone-sensitive2 lipase like este5 from a metagenome library
74 d2pbla1
not modelled
99.6
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase75 c3bjrA_
not modelled
99.6
11
PDB header: hydrolaseChain: A: PDB Molecule: putative carboxylesterase;PDBTitle: crystal structure of a putative carboxylesterase (lp_1002) from2 lactobacillus plantarum wcfs1 at 2.09 a resolution
76 c2c7bA_
not modelled
99.6
18
PDB header: hydrolaseChain: A: PDB Molecule: carboxylesterase;PDBTitle: the crystal structure of este1, a new thermophilic and2 thermostable carboxylesterase cloned from a metagenomic3 library
77 c3llcA_
not modelled
99.6
14
PDB header: hydrolaseChain: A: PDB Molecule: putative hydrolase;PDBTitle: crystal structure of putative hydrolase (yp_002548124.1) from2 agrobacterium vitis s4 at 1.80 a resolution
78 d1jfra_
not modelled
99.6
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Lipase79 c3qm1A_
not modelled
99.6
13
PDB header: hydrolaseChain: A: PDB Molecule: cinnamoyl esterase;PDBTitle: crystal structure of the lactobacillus johnsonii cinnamoyl esterase2 lj0536 s106a mutant in complex with ethylferulate, form ii
80 d1vkha_
not modelled
99.6
10
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Putative serine hydrolase Ydr428c81 d1u4na_
not modelled
99.6
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase82 c3h04A_
not modelled
99.6
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of the protein with unknown function from2 staphylococcus aureus subsp. aureus mu50
83 d1wb4a1
not modelled
99.6
9
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase84 d1jjia_
not modelled
99.6
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase85 c2wirB_
not modelled
99.6
19
PDB header: hydrolaseChain: B: PDB Molecule: alpha/beta hydrolase fold-3 domain protein;PDBTitle: hyperthermophilic esterase from the archeon pyrobaculum2 calidifontis
86 c2qruA_
not modelled
99.6
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an alpha/beta hydrolase superfamily protein from2 enterococcus faecalis
87 c3c8dA_
not modelled
99.6
15
PDB header: hydrolaseChain: A: PDB Molecule: enterochelin esterase;PDBTitle: crystal structure of the enterobactin esterase fes from2 shigella flexneri in the presence of 2,3-di-hydroxy-n-3 benzoyl-glycine
88 c2hdwB_
not modelled
99.5
14
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical protein pa2218;PDBTitle: crystal structure of hypothetical protein pa2218 from pseudomonas2 aeruginosa
89 d1ufoa_
not modelled
99.5
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Hypothetical protein TT166290 c3aikB_
not modelled
99.5
11
PDB header: hydrolaseChain: B: PDB Molecule: 303aa long hypothetical esterase;PDBTitle: crystal structure of a hsl-like carboxylesterase from sulfolobus2 tokodaii
91 c2q0xA_
not modelled
99.5
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: alpha/beta hydrolase fold protein of unknown function
92 c2gopB_
not modelled
99.5
15
PDB header: hydrolaseChain: B: PDB Molecule: trilobed protease;PDBTitle: the beta-propeller domain of the trilobed protease from pyrococcus2 furiosus reveals an open velcro topology
93 d1r88a_
not modelled
99.5
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Mycobacterial antigens94 c2uz0B_
not modelled
99.5
12
PDB header: hydrolaseChain: B: PDB Molecule: tributyrin esterase;PDBTitle: the crystal crystal structure of the esta protein, a2 virulence factor esta protein from streptococcus pneumonia
95 d1hlga_
not modelled
99.5
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Gastric lipase96 c3g8yA_
not modelled
99.5
12
PDB header: hydrolaseChain: A: PDB Molecule: susd/ragb-associated esterase-like protein;PDBTitle: crystal structure of a putative hydrolase (bvu_4111) from bacteroides2 vulgatus atcc 8482 at 1.90 a resolution
97 c2fx5A_
not modelled
99.5
17
PDB header: hydrolaseChain: A: PDB Molecule: lipase;PDBTitle: pseudomonas mendocina lipase
98 d3c8da2
not modelled
99.5
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase99 c2h1iA_
not modelled
99.5
12
PDB header: hydrolaseChain: A: PDB Molecule: carboxylesterase;PDBTitle: crystal structure of the bacillus cereus carboxylesterase
100 d1fj2a_
not modelled
99.5
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase/thioesterase 1101 c1wb4A_
not modelled
99.5
9
PDB header: hydrolaseChain: A: PDB Molecule: endo-1,4-beta-xylanase y;PDBTitle: s954a mutant of the feruloyl esterase module from2 clostridium thermocellum complexed with sinapinate
102 d2h1ia1
not modelled
99.4
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase/thioesterase 1103 d1k8qa_
not modelled
99.4
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Gastric lipase104 c3u0vA_
not modelled
99.4
13
PDB header: hydrolaseChain: A: PDB Molecule: lysophospholipase-like protein 1;PDBTitle: crystal structure analysis of human lyplal1
105 c3jw8A_
not modelled
99.4
15
PDB header: hydrolaseChain: A: PDB Molecule: mgll protein;PDBTitle: crystal structure of human mono-glyceride lipase
106 c3nuzF_
not modelled
99.4
12
PDB header: hydrolaseChain: F: PDB Molecule: putative acetyl xylan esterase;PDBTitle: crystal structure of a putative acetyl xylan esterase (bf1801) from2 bacteroides fragilis nctc 9343 at 2.30 a resolution
107 d3b5ea1
not modelled
99.4
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase/thioesterase 1108 c3hjuB_
not modelled
99.4
15
PDB header: hydrolaseChain: B: PDB Molecule: monoglyceride lipase;PDBTitle: crystal structure of human monoglyceride lipase
109 c2w8bB_
not modelled
99.4
8
PDB header: protein transport/membrane proteinChain: B: PDB Molecule: protein tolb;PDBTitle: crystal structure of processed tolb in complex with pal
110 c3og9A_
not modelled
99.4
15
PDB header: hydrolaseChain: A: PDB Molecule: protein yahd a copper inducible hydrolase;PDBTitle: structure of yahd with malic acid
111 c3dyvA_
not modelled
99.4
13
PDB header: hydrolaseChain: A: PDB Molecule: esterase d;PDBTitle: snapshots of esterase d from lactobacillus rhamnosus:2 insights into a rotation driven catalytic mechanism
112 c3cn9B_
not modelled
99.4
16
PDB header: hydrolaseChain: B: PDB Molecule: carboxylesterase;PDBTitle: crystal structure analysis of the carboxylesterase pa3859 from2 pseudomonas aeruginosa pao1- orthorhombic crystal form
113 d2bgra1
not modelled
99.3
10
Fold: 8-bladed beta-propellerSuperfamily: DPP6 N-terminal domain-likeFamily: DPP6 N-terminal domain-like114 c3picB_
not modelled
99.3
16
PDB header: hydrolaseChain: B: PDB Molecule: cip2;PDBTitle: glucuronoyl esterase catalytic domain (cip2_ge) from hypocrea jecorina
115 d1k32a2
not modelled
99.3
9
Fold: 6-bladed beta-propellerSuperfamily: Tricorn protease N-terminal domainFamily: Tricorn protease N-terminal domain116 d1orva1
not modelled
99.3
11
Fold: 8-bladed beta-propellerSuperfamily: DPP6 N-terminal domain-likeFamily: DPP6 N-terminal domain-like117 d2d81a1
not modelled
99.3
13
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: PHB depolymerase-like118 c3d59B_
not modelled
99.3
21
PDB header: hydrolaseChain: B: PDB Molecule: platelet-activating factor acetylhydrolase;PDBTitle: crystal structure of human plasma platelet activating2 factor acetylhydrolase
119 c3pe7A_
not modelled
99.3
11
PDB header: lyaseChain: A: PDB Molecule: oligogalacturonate lyase;PDBTitle: oligogalacturonate lyase in complex with manganese
120 d2b61a1
not modelled
99.3
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: O-acetyltransferase