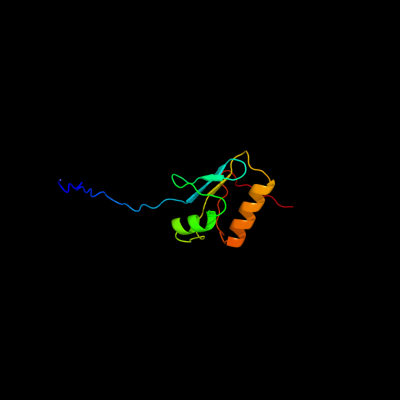
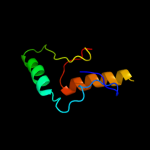
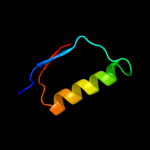
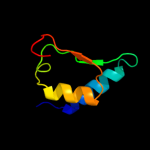
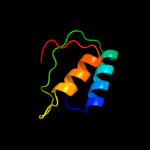
1 c2pfuA_
99.8
100
PDB header: transport proteinChain: A: PDB Molecule: biopolymer transport exbd protein;PDBTitle: nmr strcuture determination of the periplasmic domain of exbd from2 e.coli
2 c2jwlB_
99.4
28
PDB header: membrane proteinChain: B: PDB Molecule: protein tolr;PDBTitle: solution structure of periplasmic domain of tolr from h.2 influenzae with saxs data
3 d2axtk1
49.0
31
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein K, PsbKFamily: PsbK-like4 c3a0bK_
45.2
31
PDB header: electron transportChain: K: PDB Molecule: photosystem ii reaction center protein k;PDBTitle: crystal structure of br-substituted photosystem ii complex
5 c3e6eC_
35.6
10
PDB header: isomeraseChain: C: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from e.faecalis2 complex with cycloserine
6 c3a0bk_
35.2
31
PDB header: electron transportChain: K: PDB Molecule: photosystem ii reaction center protein k;PDBTitle: crystal structure of br-substituted photosystem ii complex
7 d1bd0a2
29.5
19
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain8 c2jvfA_
28.2
21
PDB header: de novo proteinChain: A: PDB Molecule: de novo protein m7;PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
9 c2re3A_
27.6
13
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf1285 family protein (spo_0140) from2 silicibacter pomeroyi dss-3 at 2.50 a resolution
10 c3a0hk_
26.0
31
PDB header: electron transportChain: K: PDB Molecule: photosystem ii reaction center protein k;PDBTitle: crystal structure of i-substituted photosystem ii complex
11 d2e8aa1
25.1
18
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP7012 d1jcea1
25.1
16
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP7013 c1qysA_
22.5
17
PDB header: de novo proteinChain: A: PDB Molecule: top7;PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
14 d1dkgd1
20.9
14
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP7015 d1bupa1
19.8
18
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP7016 d1vfsa2
19.8
16
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain17 c1ur8B_
18.4
12
PDB header: hydrolaseChain: B: PDB Molecule: chitinase b;PDBTitle: interactions of a family 18 chitinase with the designed2 inhibitor hm508, and its degradation product,3 chitobiono-delta-lactone
18 c3ugsB_
18.3
11
PDB header: transferaseChain: B: PDB Molecule: undecaprenyl pyrophosphate synthase;PDBTitle: crystal structure of a probable undecaprenyl diphosphate synthase2 (upps) from campylobacter jejuni
19 c1vftA_
17.8
16
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: crystal structure of l-cycloserine-bound form of alanine2 racemase from d-cycloserine-producing streptomyces3 lavendulae
20 c3ghfA_
17.5
11
PDB header: cell cycleChain: A: PDB Molecule: septum site-determining protein minc;PDBTitle: crystal structure of the septum site-determining protein2 minc from salmonella typhimurium
21 c3co8B_
not modelled
16.6
12
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from oenococcus oeni
22 c3lupA_
not modelled
16.5
18
PDB header: structure genomics, unknown functionChain: A: PDB Molecule: degv family protein;PDBTitle: crystal structure of fatty acid binding degv family protein sag13422 from streptococcus agalactiae
23 c2c2xB_
not modelled
16.3
9
PDB header: oxidoreductaseChain: B: PDB Molecule: methylenetetrahydrofolate dehydrogenase-PDBTitle: three dimensional structure of bifunctional2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase3 from mycobacterium tuberculosis
24 d1edza2
not modelled
16.1
9
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Tetrahydrofolate dehydrogenase/cyclohydrolase25 c3me8B_
not modelled
15.8
11
PDB header: electron transportChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of putative electron transfer protein aq_2194 from2 aquifex aeolicus vf5
26 c1niuA_
not modelled
15.7
19
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: alanine racemase with bound inhibitor derived from l-2 cycloserine
27 d1qyia_
not modelled
14.9
15
Fold: HAD-likeSuperfamily: HAD-likeFamily: Hypothetical protein MW1667 (SA1546)28 d1goia2
not modelled
14.8
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase29 c3oo2A_
not modelled
13.6
12
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase 1;PDBTitle: 2.37 angstrom resolution crystal structure of an alanine racemase2 (alr) from staphylococcus aureus subsp. aureus col
30 d2d0oa1
not modelled
12.5
17
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Swiveling domain of dehydratase reactivase alpha subunitFamily: Swiveling domain of dehydratase reactivase alpha subunit31 c3pm6B_
not modelled
12.3
14
PDB header: lyaseChain: B: PDB Molecule: putative fructose-bisphosphate aldolase;PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
32 c3outC_
not modelled
11.9
15
PDB header: isomeraseChain: C: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase from francisella tularensis2 subsp. tularensis schu s4 in complex with d-glutamate.
33 d1a9xa4
not modelled
11.7
11
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like34 d2j9ga2
not modelled
11.5
7
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like35 c2g7zB_
not modelled
11.4
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein spy1493;PDBTitle: conserved degv-like protein of unknown function from streptococcus2 pyogenes m1 gas binds long-chain fatty acids
36 c3iprC_
not modelled
10.8
17
PDB header: transferaseChain: C: PDB Molecule: pts system, iia component;PDBTitle: crystal structure of the enterococcus faecalis gluconate2 specific eiia phosphotransferase system component
37 d1b0aa2
not modelled
10.8
4
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Tetrahydrofolate dehydrogenase/cyclohydrolase38 d1wd5a_
not modelled
10.8
11
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)39 c1xfcB_
not modelled
10.7
19
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: the 1.9 a crystal structure of alanine racemase from mycobacterium2 tuberculosis contains a conserved entryway into the active site
40 d2axth1
not modelled
10.5
18
Fold: Single transmembrane helixSuperfamily: Photosystem II 10 kDa phosphoprotein PsbHFamily: PsbH-like41 d1ne9a1
not modelled
10.5
18
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: FemXAB nonribosomal peptidyltransferases42 d2p10a1
not modelled
10.0
8
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Mll9387-like43 c2dy3B_
not modelled
9.9
14
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from corynebacterium glutamicum
44 c2ra9A_
not modelled
9.5
5
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein duf1285;PDBTitle: crystal structure of a duf1285 family protein (sbal_2486) from2 shewanella baltica os155 at 1.40 a resolution
45 c2odoC_
not modelled
9.4
15
PDB header: isomeraseChain: C: PDB Molecule: alanine racemase;PDBTitle: crystal structure of pseudomonas fluorescens alanine racemase
46 d1f75a_
not modelled
9.2
19
Fold: Undecaprenyl diphosphate synthaseSuperfamily: Undecaprenyl diphosphate synthaseFamily: Undecaprenyl diphosphate synthase47 c1p74B_
not modelled
9.2
21
PDB header: oxidoreductaseChain: B: PDB Molecule: shikimate 5-dehydrogenase;PDBTitle: crystal structure of shikimate dehydrogenase (aroe) from2 haemophilus influenzae
48 c3a0hY_
not modelled
8.7
10
PDB header: electron transportChain: Y: PDB Molecule: photosystem ii reaction center protein ycf12;PDBTitle: crystal structure of i-substituted photosystem ii complex
49 c3bz1y_
not modelled
8.7
10
PDB header: electron transportChain: Y: PDB Molecule: photosystem ii protein y;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
50 c3a0hy_
not modelled
8.7
10
PDB header: electron transportChain: Y: PDB Molecule: photosystem ii reaction center protein ycf12;PDBTitle: crystal structure of i-substituted photosystem ii complex
51 c3a0by_
not modelled
8.5
10
PDB header: electron transportChain: Y: PDB Molecule: photosystem ii reaction center protein ycf12;PDBTitle: crystal structure of br-substituted photosystem ii complex
52 d1gph11
not modelled
8.4
15
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)53 c3arcY_
not modelled
8.3
10
PDB header: electron transport, photosynthesisChain: Y: PDB Molecule: protein ycf12;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
54 c3arcy_
not modelled
8.3
10
PDB header: electron transport, photosynthesisChain: Y: PDB Molecule: protein ycf12;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
55 d1gvfa_
not modelled
8.3
19
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class II FBP aldolase56 c1jp3A_
not modelled
8.0
10
PDB header: transferaseChain: A: PDB Molecule: undecaprenyl pyrophosphate synthase;PDBTitle: structure of e.coli undecaprenyl pyrophosphate synthase
57 d1ecfa1
not modelled
7.9
20
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)58 c3eooL_
not modelled
7.8
15
PDB header: lyaseChain: L: PDB Molecule: methylisocitrate lyase;PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
59 d2nn6d2
not modelled
7.7
8
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like60 d1hgxa_
not modelled
7.6
8
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)61 d1rvga_
not modelled
7.5
13
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class II FBP aldolase62 c3qm3C_
not modelled
7.4
9
PDB header: lyaseChain: C: PDB Molecule: fructose-bisphosphate aldolase;PDBTitle: 1.85 angstrom resolution crystal structure of fructose-bisphosphate2 aldolase (fba) from campylobacter jejuni
63 c3gmgB_
not modelled
7.3
26
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein rv1825/mt1873;PDBTitle: crystal structure of an uncharacterized conserved protein2 from mycobacterium tuberculosis
64 c3o8qB_
not modelled
7.2
21
PDB header: oxidoreductaseChain: B: PDB Molecule: shikimate 5-dehydrogenase i alpha;PDBTitle: 1.45 angstrom resolution crystal structure of shikimate 5-2 dehydrogenase (aroe) from vibrio cholerae
65 c2vfwB_
not modelled
6.8
14
PDB header: transferaseChain: B: PDB Molecule: short-chain z-isoprenyl diphosphate synthetase;PDBTitle: rv1086 native
66 d1ueha_
not modelled
6.8
10
Fold: Undecaprenyl diphosphate synthaseSuperfamily: Undecaprenyl diphosphate synthaseFamily: Undecaprenyl diphosphate synthase67 d1zhva2
not modelled
6.7
37
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Atu0741-like68 c3gycB_
not modelled
6.7
32
PDB header: hydrolaseChain: B: PDB Molecule: putative glycoside hydrolase;PDBTitle: crystal structure of putative glycoside hydrolase (yp_001304622.1)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
69 c3a0bY_
not modelled
6.7
10
PDB header: electron transportChain: Y: PDB Molecule: photosystem ii reaction center protein ycf12;PDBTitle: crystal structure of br-substituted photosystem ii complex
70 d1g8fa3
not modelled
6.7
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ATP sulfurylase C-terminal domain71 c2d2rA_
not modelled
6.2
14
PDB header: transferaseChain: A: PDB Molecule: undecaprenyl pyrophosphate synthase;PDBTitle: crystal structure of helicobacter pylori undecaprenyl pyrophosphate2 synthase
72 c2f40A_
not modelled
6.0
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pf1455;PDBTitle: structure of a novel protein from backbone-centered nmr data and nmr-2 assisted structure prediction
73 d1i5ea_
not modelled
5.8
11
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)74 c2qlxA_
not modelled
5.8
31
PDB header: isomeraseChain: A: PDB Molecule: l-rhamnose mutarotase;PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum in complex with l-rhamnose
75 c2qlwA_
not modelled
5.8
31
PDB header: isomeraseChain: A: PDB Molecule: rhau;PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum
76 c2dcqA_
not modelled
5.8
9
PDB header: unknown functionChain: A: PDB Molecule: putative protein at4g01050;PDBTitle: fully automated nmr structure determination of the2 rhodanese homology domain at4g01050(175-295) from3 arabidopsis thaliana
77 c3oo2B_
not modelled
5.7
16
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase 1;PDBTitle: 2.37 angstrom resolution crystal structure of an alanine racemase2 (alr) from staphylococcus aureus subsp. aureus col
78 d1zzoa1
not modelled
5.7
16
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like79 d2bz1a1
not modelled
5.5
20
Fold: RibA-likeSuperfamily: RibA-likeFamily: RibA-like80 d1x8da1
not modelled
5.5
13
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: YiiL-like81 c1ecjB_
not modelled
5.3
20
PDB header: transferaseChain: B: PDB Molecule: glutamine phosphoribosylpyrophosphatePDBTitle: escherichia coli glutamine phosphoribosylpyrophosphate2 (prpp) amidotransferase complexed with 2 amp per tetramer
82 d1g9sa_
not modelled
5.2
16
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)83 c1yfzA_
not modelled
5.2
14
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: novel imp binding in feedback inhibition of hypoxanthine-guanine2 phosphoribosyltransferase from thermoanaerobacter tengcongensis
84 d1yfza1
not modelled
5.2
14
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)85 c3l8cA_
not modelled
5.2
15
PDB header: ligaseChain: A: PDB Molecule: d-alanine--poly(phosphoribitol) ligase subunit 1;PDBTitle: structure of probable d-alanine--poly(phosphoribitol) ligase2 subunit-1 from streptococcus pyogenes
86 c3q94B_
not modelled
5.2
12
PDB header: lyaseChain: B: PDB Molecule: fructose-bisphosphate aldolase, class ii;PDBTitle: the crystal structure of fructose 1,6-bisphosphate aldolase from2 bacillus anthracis str. 'ames ancestor'
87 c3navB_
not modelled
5.1
15
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
88 d1xd7a_
not modelled
5.1
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcriptional regulator Rrf289 d1aopa1
not modelled
5.1
15
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 390 d1ujpa_
not modelled
5.1
9
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes91 d1ulza2
not modelled
5.1
14
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like92 d1rhsa1
not modelled
5.1
10
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)93 d1zj8a1
not modelled
5.0
14
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 3