| 1 | c2kxoA_
|
|
 |
100.0 |
42 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division topological specificity factor;
PDBTitle: solution nmr structure of the cell division regulator mine protein2 from neisseria gonorrhoeae
|
| 2 | c3r9jD_
|
|
 |
99.9 |
99 |
PDB header:cell cycle,hydrolase/cell cycle
Chain: D: PDB Molecule:cell division topological specificity factor;
PDBTitle: 4.3a resolution structure of a mind-mine(i24n) protein complex
|
| 3 | d1ev0a_
|
|
 |
99.6 |
100 |
Fold:Cell division protein MinE topological specificity domain
Superfamily:Cell division protein MinE topological specificity domain
Family:Cell division protein MinE topological specificity domain |
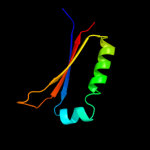
| 4 | c3ku7B_
|
|
 |
98.8 |
33 |
PDB header:cell cycle
Chain: B: PDB Molecule:cell division topological specificity factor;
PDBTitle: crystal structure of helicobacter pylori mine, a cell division2 topological specificity factor
|
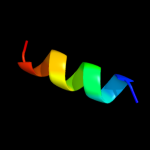
| 5 | c3r9iG_
|
|
 |
88.6 |
100 |
PDB header:cell cycle,hydrolase/cell cycle
Chain: G: PDB Molecule:cell division topological specificity factor;
PDBTitle: 2.6a resolution structure of mind complexed with mine (12-31) peptide
|
| 6 | c3r9iH_
|
|
 |
85.9 |
100 |
PDB header:cell cycle,hydrolase/cell cycle
Chain: H: PDB Molecule:cell division topological specificity factor;
PDBTitle: 2.6a resolution structure of mind complexed with mine (12-31) peptide
|
| 7 | c3r9iE_
|
|
 |
85.9 |
100 |
PDB header:cell cycle,hydrolase/cell cycle
Chain: E: PDB Molecule:cell division topological specificity factor;
PDBTitle: 2.6a resolution structure of mind complexed with mine (12-31) peptide
|
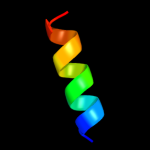
| 8 | c3r9iF_
|
|
 |
85.9 |
100 |
PDB header:cell cycle,hydrolase/cell cycle
Chain: F: PDB Molecule:cell division topological specificity factor;
PDBTitle: 2.6a resolution structure of mind complexed with mine (12-31) peptide
|
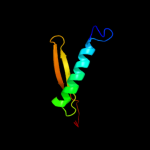
| 9 | c2kwpA_
|
|
 |
75.9 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation protein nusa;
PDBTitle: solution structure of the aminoterminal domain of e. coli nusa
|
| 10 | d1hh2p4
|
|
 |
74.7 |
21 |
Fold:Transcription factor NusA, N-terminal domain
Superfamily:Transcription factor NusA, N-terminal domain
Family:Transcription factor NusA, N-terminal domain |
| 11 | c3epsB_
|
|
 |
55.7 |
22 |
PDB header:transferase, hydrolase
Chain: B: PDB Molecule:isocitrate dehydrogenase kinase/phosphatase;
PDBTitle: the crystal structure of isocitrate dehydrogenase kinase/phosphatase2 from e. coli
|
| 12 | d2r40d1
|
|
 |
36.5 |
16 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 13 | d1vmaa1
|
|
 |
33.7 |
16 |
Fold:Four-helical up-and-down bundle
Superfamily:Domain of the SRP/SRP receptor G-proteins
Family:Domain of the SRP/SRP receptor G-proteins |
| 14 | c1rwuA_
|
|
 |
31.6 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical upf0250 protein ybed;
PDBTitle: solution structure of conserved protein ybed from e. coli
|
| 15 | d1rwua_
|
|
 |
31.6 |
20 |
Fold:Ferredoxin-like
Superfamily:YbeD/HP0495-like
Family:YbeD-like |
| 16 | d2qw4a1
|
|
 |
31.4 |
19 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 17 | c2nxxG_
|
|
 |
30.9 |
22 |
PDB header:hormone/growth factor
Chain: G: PDB Molecule:ecdysone receptor (ecr, nrh1);
PDBTitle: crystal structure of the ligand-binding domains of the2 t.castaneum (coleoptera) heterodimer ecrusp bound to3 ponasterone a
|
| 18 | c3ctbA_
|
|
 |
28.7 |
16 |
PDB header:transcription, transferase
Chain: A: PDB Molecule:pregnane x receptor, linker, steroid receptor
PDBTitle: tethered pxr-lbd/src-1p apoprotein
|
| 19 | d1ovle_
|
|
 |
22.5 |
19 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 20 | c1nrlA_
|
|
 |
21.9 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:orphan nuclear receptor pxr;
PDBTitle: crystal structure of the human pxr-lbd in complex with an2 src-1 coactivator peptide and sr12813
|
| 21 | d1nrla_ |
|
not modelled |
21.9 |
16 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 22 | d1nq7a_ |
|
not modelled |
21.6 |
19 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 23 | c3hm5A_ |
|
not modelled |
21.4 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:dna methyltransferase 1-associated protein 1;
PDBTitle: sant domain of human dna methyltransferase 1 associated2 protein 1
|
| 24 | d1d0qa_ |
|
not modelled |
20.2 |
32 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:DNA primase zinc finger |
| 25 | c1z5xE_ |
|
not modelled |
20.1 |
22 |
PDB header:hormone/growth factor receptor
Chain: E: PDB Molecule:ecdysone receptor ligand binding domain;
PDBTitle: hemipteran ecdysone receptor ligand-binding domain2 complexed with ponasterone a
|
| 26 | c3pvpA_ |
|
not modelled |
20.0 |
17 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:chromosomal replication initiator protein dnaa;
PDBTitle: structure of mycobacterium tuberculosis dnaa-dbd in complex with box22 dna
|
| 27 | d1j1va_ |
|
not modelled |
19.5 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:TrpR-like
Family:Chromosomal replication initiation factor DnaA C-terminal domain IV |
| 28 | d1go3f_ |
|
not modelled |
19.3 |
33 |
Fold:SAM domain-like
Superfamily:HRDC-like
Family:RNA polymerase II subunit RBP4 (RpoF) |
| 29 | c2dnwA_ |
|
not modelled |
18.4 |
32 |
PDB header:transport protein
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: solution structure of rsgi ruh-059, an acp domain of acyl2 carrier protein, mitochondrial [precursor] from human cdna
|
| 30 | d1ovla_ |
|
not modelled |
17.8 |
19 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 31 | d1pdua_ |
|
not modelled |
17.6 |
22 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 32 | d1n83a_ |
|
not modelled |
17.4 |
16 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 33 | c2qw4D_ |
|
not modelled |
17.3 |
19 |
PDB header:hormone receptor
Chain: D: PDB Molecule:orphan nuclear receptor nr4a1;
PDBTitle: human nr4a1 ligand-binding domain
|
| 34 | c1xdkF_ |
|
not modelled |
16.1 |
22 |
PDB header:hormone/growth factor receptor
Chain: F: PDB Molecule:retinoic acid receptor, beta;
PDBTitle: crystal structure of the rarbeta/rxralpha ligand binding2 domain heterodimer in complex with 9-cis retinoic acid and3 a fragment of the trap220 coactivator
|
| 35 | d1pq9a_ |
|
not modelled |
15.8 |
22 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 36 | d1osva_ |
|
not modelled |
15.1 |
19 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 37 | d1nava_ |
|
not modelled |
13.6 |
13 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 38 | d1l8qa1 |
|
not modelled |
13.6 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:TrpR-like
Family:Chromosomal replication initiation factor DnaA C-terminal domain IV |
| 39 | c3up0A_ |
|
not modelled |
13.1 |
6 |
PDB header:steroid binding protein/transcription
Chain: A: PDB Molecule:acedaf-12;
PDBTitle: nuclear receptor daf-12 from hookworm ancylostoma ceylanicum in2 complex with (25s)-delta7-dafachronic acid
|
| 40 | c3pt8B_ |
|
not modelled |
13.1 |
7 |
PDB header:oxygen transport
Chain: B: PDB Molecule:hemoglobin iii;
PDBTitle: structure of hbii-iii-cn from lucina pectinata at ph 5.0
|
| 41 | c2c0kB_ |
|
not modelled |
13.0 |
19 |
PDB header:oxygen transport
Chain: B: PDB Molecule:hemoglobin;
PDBTitle: the structure of hemoglobin from the botfly gasterophilus2 intestinalis
|
| 42 | c3l0jA_ |
|
not modelled |
12.6 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:nuclear receptor ror-gamma;
PDBTitle: crystal structure of orphan nuclear receptor rorgamma in complex with2 natural ligand
|
| 43 | c2rhfA_ |
|
not modelled |
12.4 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:dna helicase recq;
PDBTitle: d. radiodurans recq hrdc domain 3
|
| 44 | d1ie9a_ |
|
not modelled |
12.4 |
13 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 45 | c2hc4A_ |
|
not modelled |
12.1 |
13 |
PDB header:gene regulation
Chain: A: PDB Molecule:vitamin d receptor;
PDBTitle: crystal structure of the lbd of vdr of danio rerio in2 complex with calcitriol
|
| 46 | d1ghha_ |
|
not modelled |
11.6 |
18 |
Fold:DNA damage-inducible protein DinI
Superfamily:DNA damage-inducible protein DinI
Family:DNA damage-inducible protein DinI |
| 47 | d1sqna_ |
|
not modelled |
11.2 |
6 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 48 | c2oaxC_ |
|
not modelled |
10.7 |
10 |
PDB header:transcription
Chain: C: PDB Molecule:mineralocorticoid receptor;
PDBTitle: crystal structure of the s810l mutant mineralocorticoid2 receptor associated with sc9420
|
| 49 | c3lbdA_ |
|
not modelled |
10.6 |
22 |
PDB header:nuclear receptor
Chain: A: PDB Molecule:retinoic acid receptor gamma;
PDBTitle: ligand-binding domain of the human retinoic acid receptor2 gamma bound to 9-cis retinoic acid
|
| 50 | d1x9fb_ |
|
not modelled |
10.5 |
21 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 51 | c3vhuA_ |
|
not modelled |
10.4 |
10 |
PDB header:transcription/inhibitor
Chain: A: PDB Molecule:mineralocorticoid receptor;
PDBTitle: mineralocorticoid receptor ligand-binding domain with spironolactone
|
| 52 | c1p93A_ |
|
not modelled |
10.4 |
6 |
PDB header:hormone receptor
Chain: A: PDB Molecule:glucocorticoid receptor;
PDBTitle: crystal structure of the agonist form of glucocorticoid2 receptor
|
| 53 | c2o4rA_ |
|
not modelled |
10.0 |
13 |
PDB header:hormone/growth factor receptor
Chain: A: PDB Molecule:vitamin d3 receptor;
PDBTitle: crystal structure of rat vitamin d receptor ligand binding2 domain complexed with vitiii 17-20e and the nr2 box of3 drip 205
|
| 54 | c3gdzA_ |
|
not modelled |
9.5 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:arginyl-trna synthetase;
PDBTitle: crystal structure of arginyl-trna synthetase from klebsiella2 pneumoniae subsp. pneumoniae
|
| 55 | d1xapa_ |
|
not modelled |
8.9 |
22 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 56 | d1xvpb_ |
|
not modelled |
8.8 |
13 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 57 | d3proc1 |
|
not modelled |
8.6 |
18 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Alpha-lytic protease prodomain
Family:Alpha-lytic protease prodomain |
| 58 | d1itha_ |
|
not modelled |
8.5 |
23 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 59 | c2olpB_ |
|
not modelled |
8.5 |
5 |
PDB header:oxygen storage/transport
Chain: B: PDB Molecule:hemoglobin ii;
PDBTitle: structure and ligand selection of hemoglobin ii from lucina pectinata
|
| 60 | c3ifeA_ |
|
not modelled |
8.4 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidase t;
PDBTitle: 1.55 angstrom resolution crystal structure of peptidase t (pept-1)2 from bacillus anthracis str. 'ames ancestor'.
|
| 61 | c3abfB_ |
|
not modelled |
8.2 |
21 |
PDB header:isomerase
Chain: B: PDB Molecule:4-oxalocrotonate tautomerase;
PDBTitle: crystal structure of a 4-oxalocrotonate tautomerase homologue2 (tthb242)
|
| 62 | d2zmia1 |
|
not modelled |
8.2 |
13 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 63 | d2b50a1 |
|
not modelled |
8.2 |
15 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 64 | c3q9qB_ |
|
not modelled |
7.8 |
29 |
PDB header:chaperone
Chain: B: PDB Molecule:heat shock protein beta-1;
PDBTitle: hspb1 fragment second crystal form
|
| 65 | d1mbaa_ |
|
not modelled |
7.7 |
9 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 66 | d2ia7a1 |
|
not modelled |
7.4 |
11 |
Fold:gpW/gp25-like
Superfamily:gpW/gp25-like
Family:gpW/gp25-like |
| 67 | d2dgxa1 |
|
not modelled |
7.1 |
15 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 68 | c2x4kB_ |
|
not modelled |
7.0 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:4-oxalocrotonate tautomerase;
PDBTitle: crystal structure of sar1376, a putative 4-oxalocrotonate2 tautomerase from the methicillin-resistant staphylococcus3 aureus (mrsa)
|
| 69 | c1usdA_ |
|
not modelled |
6.9 |
21 |
PDB header:signaling protein
Chain: A: PDB Molecule:vasodilator-stimulated phosphoprotein;
PDBTitle: human vasp tetramerisation domain l352m
|
| 70 | c2rqlA_ |
|
not modelled |
6.7 |
8 |
PDB header:translation
Chain: A: PDB Molecule:probable sigma-54 modulation protein;
PDBTitle: solution structure of the e. coli ribosome hibernation2 promoting factor hpf
|
| 71 | c3mb2G_ |
|
not modelled |
6.7 |
6 |
PDB header:isomerase
Chain: G: PDB Molecule:4-oxalocrotonate tautomerase family enzyme - alpha subunit;
PDBTitle: kinetic and structural characterization of a heterohexamer 4-2 oxalocrotonate tautomerase from chloroflexus aurantiacus j-10-fl:3 implications for functional and structural diversity in the4 tautomerase superfamily
|
| 72 | d1bjpa_ |
|
not modelled |
6.4 |
15 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:4-oxalocrotonate tautomerase-like |
| 73 | d1wuda1 |
|
not modelled |
6.3 |
21 |
Fold:SAM domain-like
Superfamily:HRDC-like
Family:HRDC domain from helicases |
| 74 | d1lhta_ |
|
not modelled |
6.2 |
20 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 75 | d1vkua_ |
|
not modelled |
6.2 |
22 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Acyl-carrier protein (ACP) |
| 76 | c3idwA_ |
|
not modelled |
6.2 |
15 |
PDB header:endocytosis
Chain: A: PDB Molecule:actin cytoskeleton-regulatory complex protein sla1;
PDBTitle: crystal structure of sla1 homology domain 2
|
| 77 | c2zs0A_ |
|
not modelled |
6.1 |
11 |
PDB header:oxygen storage, oxygen transport
Chain: A: PDB Molecule:extracellular giant hemoglobin major globin subunit a1;
PDBTitle: structural basis for the heterotropic and homotropic interactions of2 invertebrate giant hemoglobin
|
| 78 | c2iz2A_ |
|
not modelled |
6.1 |
9 |
PDB header:dna binding protein
Chain: A: PDB Molecule:nuclear hormone receptor ftz-f1;
PDBTitle: crystal structure of the ligand binding domain of fushi2 tarazu factor 1 from drosophila melanogaster
|
| 79 | d2fvja1 |
|
not modelled |
6.0 |
11 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 80 | d1oj6a_ |
|
not modelled |
5.9 |
9 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 81 | c2yruA_ |
|
not modelled |
5.8 |
7 |
PDB header:apoptosis
Chain: A: PDB Molecule:steroid receptor rna activator 1;
PDBTitle: solution structure of mouse steroid receptor rna activator2 1 (sra1) protein
|
| 82 | d1d02a_ |
|
not modelled |
5.8 |
28 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Restriction endonuclease MunI |
| 83 | d2qssa1 |
|
not modelled |
5.8 |
11 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 84 | c3tqmD_ |
|
not modelled |
5.7 |
19 |
PDB header:protein binding
Chain: D: PDB Molecule:ribosome-associated factor y;
PDBTitle: structure of an ribosomal subunit interface protein from coxiella2 burnetii
|
| 85 | c2ehtA_ |
|
not modelled |
5.5 |
24 |
PDB header:lipid transport
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: crystal structure of acyl carrier protein from aquifex aeolicus (form2 2)
|
| 86 | d1sctb_ |
|
not modelled |
5.5 |
10 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 87 | d1l4sa_ |
|
not modelled |
5.5 |
10 |
Fold:Ribosome binding protein Y (YfiA homologue)
Superfamily:Ribosome binding protein Y (YfiA homologue)
Family:Ribosome binding protein Y (YfiA homologue) |
| 88 | c2op8A_ |
|
not modelled |
5.4 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:probable tautomerase ywhb;
PDBTitle: crystal structure of ywhb- homologue of 4-oxalocrotonate tautomerase
|
| 89 | d1t7ra_ |
|
not modelled |
5.4 |
6 |
Fold:Nuclear receptor ligand-binding domain
Superfamily:Nuclear receptor ligand-binding domain
Family:Nuclear receptor ligand-binding domain |
| 90 | c2qnwA_ |
|
not modelled |
5.3 |
13 |
PDB header:signaling protein
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: toxoplasma gondii apicoplast-targeted acyl carrier protein
|
| 91 | d2qssb1 |
|
not modelled |
5.3 |
6 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 92 | c3gyuA_ |
|
not modelled |
5.2 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:nuclear hormone receptor of the steroid/thyroid
PDBTitle: nuclear receptor daf-12 from parasitic nematode2 strongyloides stercoralis in complex with its physiological3 ligand dafachronic acid delta 7
|
| 93 | d1v4wb_ |
|
not modelled |
5.1 |
15 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 94 | d2htja1 |
|
not modelled |
5.1 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:FaeA-like |
| 95 | d1fsla_ |
|
not modelled |
5.1 |
10 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 96 | c2zufA_ |
|
not modelled |
5.0 |
12 |
PDB header:ligase/rna
Chain: A: PDB Molecule:arginyl-trna synthetase;
PDBTitle: crystal structure of pyrococcus horikoshii arginyl-trna2 synthetase complexed with trna(arg)
|