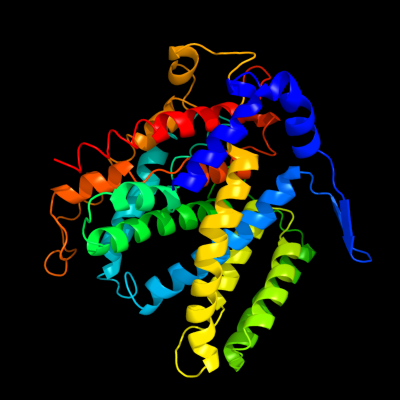
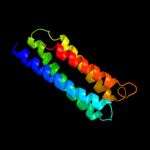
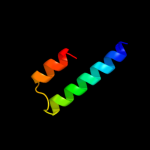
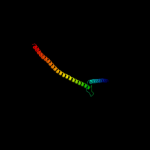
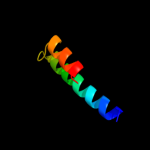
1 c1zcdA_
99.9
14
PDB header: membrane proteinChain: A: PDB Molecule: na(+)/h(+) antiporter 1;PDBTitle: crystal structure of the na+/h+ antiporter nhaa
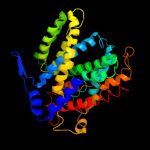
2 c2l0eA_
93.7
22
PDB header: membrane proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: structural and functional analysis of tm vi of the nhe1 isoform of the2 na+/h+ exchanger
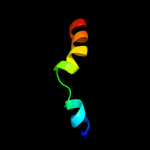
3 c2kbvA_
84.4
26
PDB header: membrane proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: structural and functional analysis of tm xi of the nhe12 isoform of the na+/h+ exchanger
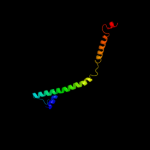
4 c3ojaB_
43.8
11
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
5 d1f6ga_
37.9
14
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels6 c3hzsA_
33.3
12
PDB header: transferaseChain: A: PDB Molecule: monofunctional glycosyltransferase;PDBTitle: s. aureus monofunctional glycosyltransferase (mtga)in complex with2 moenomycin
7 c1y4cA_
32.4
13
PDB header: de novo proteinChain: A: PDB Molecule: maltose binding protein fused with designedPDBTitle: designed helical protein fusion mbp
8 d1vqop1
29.1
19
Fold: Ribosomal protein L19 (L19e)Superfamily: Ribosomal protein L19 (L19e)Family: Ribosomal protein L19 (L19e)9 c2zv4O_
28.0
11
PDB header: structural proteinChain: O: PDB Molecule: major vault protein;PDBTitle: the structure of rat liver vault at 3.5 angstrom resolution
10 c3ojaA_
25.2
11
PDB header: protein bindingChain: A: PDB Molecule: leucine-rich immune molecule 1;PDBTitle: crystal structure of lrim1/apl1c complex
11 c3dtpA_
24.0
11
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain chimera of smooth andPDBTitle: tarantula heavy meromyosin obtained by flexible docking to2 tarantula muscle thick filament cryo-em 3d-map
12 d2csba1
22.4
14
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Topoisomerase V repeat domain13 c4a1cO_
22.3
16
PDB header: ribosomeChain: O: PDB Molecule: rpl19;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
14 c3exmA_
20.6
6
PDB header: hydrolaseChain: A: PDB Molecule: phosphatase sc4828;PDBTitle: crystal structure of the phosphatase sc4828 with the non-hydrolyzable2 nucleotide gpcp
15 c2zkrp_
20.6
19
PDB header: ribosomal protein/rnaChain: P: PDB Molecule: rna expansion segment es31 part i;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
16 d2nwwa1
19.8
19
Fold: Proton glutamate symport proteinSuperfamily: Proton glutamate symport proteinFamily: Proton glutamate symport protein17 c1i84V_
19.7
15
PDB header: contractile proteinChain: V: PDB Molecule: smooth muscle myosin heavy chain;PDBTitle: cryo-em structure of the heavy meromyosin subfragment of2 chicken gizzard smooth muscle myosin with regulatory light3 chain in the dephosphorylated state. only c alphas4 provided for regulatory light chain. only backbone atoms5 provided for s2 fragment.
18 c2dfsA_
16.9
13
PDB header: contractile protein/transport proteinChain: A: PDB Molecule: myosin-5a;PDBTitle: 3-d structure of myosin-v inhibited state
19 c3iz5T_
15.7
14
PDB header: ribosomeChain: T: PDB Molecule: 60s ribosomal protein l19 (l19e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
20 c3h36A_
15.4
12
PDB header: transferaseChain: A: PDB Molecule: polyribonucleotide nucleotidyltransferase;PDBTitle: structure of an uncharacterized domain in polyribonucleotide2 nucleotidyltransferase from streptococcus mutans ua159
21 d2c0sa1
not modelled
14.0
15
Fold: ROP-likeSuperfamily: BAS1536-likeFamily: BAS1536-like22 c2v0pA_
not modelled
13.7
8
PDB header: hydrolase inhibitorChain: A: PDB Molecule: type 2a phosphatase-associated protein 42;PDBTitle: the structure of tap42 alpha4 subunit
23 c1y4eA_
not modelled
13.3
24
PDB header: membrane proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: nmr structure of transmembrane segment iv of the nhe12 isoform of the na+/h+ exchanger
24 d2oqoa1
not modelled
12.1
16
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: PBP transglycosylase domain-like25 c3rl0j_
not modelled
11.5
17
PDB header: membrane protein/exocytosisChain: J: PDB Molecule: syntaxin-1a;PDBTitle: truncated snare complex with complexin (p1)
26 d1rkla_
not modelled
11.2
21
Fold: Single transmembrane helixSuperfamily: Oligosaccharyltransferase subunit ost4pFamily: Oligosaccharyltransferase subunit ost4p27 d2bzba1
not modelled
10.5
22
Fold: ROP-likeSuperfamily: BAS1536-likeFamily: BAS1536-like28 c3layF_
not modelled
9.9
19
PDB header: metal binding proteinChain: F: PDB Molecule: zinc resistance-associated protein;PDBTitle: alpha-helical barrel formed by the decamer of the zinc resistance-2 associated protein (stm4172) from salmonella enterica subsp. enterica3 serovar typhimurium str. lt2
29 c1kilE_
not modelled
9.7
14
PDB header: membrane proteinChain: E: PDB Molecule: complexin i snare-complex binding region;PDBTitle: three-dimensional structure of the complexin/snare complex
30 d1jkva_
not modelled
9.4
24
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Manganese catalase (T-catalase)31 c2l2lA_
not modelled
9.4
32
PDB header: transferaseChain: A: PDB Molecule: transcriptional repressor p66-alpha;PDBTitle: solution structure of the coiled-coil complex between mbd2 and2 p66alpha
32 c2akfA_
not modelled
9.3
42
PDB header: protein bindingChain: A: PDB Molecule: coronin-1a;PDBTitle: crystal structure of the coiled-coil domain of coronin 1
33 c2akfC_
not modelled
9.3
42
PDB header: protein bindingChain: C: PDB Molecule: coronin-1a;PDBTitle: crystal structure of the coiled-coil domain of coronin 1
34 c2akfB_
not modelled
9.3
42
PDB header: protein bindingChain: B: PDB Molecule: coronin-1a;PDBTitle: crystal structure of the coiled-coil domain of coronin 1
35 c3f1iH_
not modelled
9.2
18
PDB header: protein bindingChain: H: PDB Molecule: hepatocyte growth factor-regulated tyrosine kinasePDBTitle: human escrt-0 core complex
36 c2kncA_
not modelled
9.1
13
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
37 c3lt7D_
not modelled
8.9
25
PDB header: cell adhesionChain: D: PDB Molecule: adhesin yada;PDBTitle: a transition from strong right-handed to canonical left-handed2 supercoiling in a conserved coiled coil segment of trimeric3 autotransporter adhesins - the m3 mutant structure
38 c1ei3C_
not modelled
8.8
9
PDB header: PDB COMPND: 39 c1nh2D_
not modelled
8.7
21
PDB header: transcription/dnaChain: D: PDB Molecule: transcription initiation factor iia small chain;PDBTitle: crystal structure of a yeast tfiia/tbp/dna complex
40 c3dinD_
not modelled
8.7
25
PDB header: membrane protein, protein transportChain: D: PDB Molecule: preprotein translocase subunit sece;PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
41 d2olua1
not modelled
8.6
16
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: PBP transglycosylase domain-like42 c2pfmA_
not modelled
8.4
16
PDB header: lyaseChain: A: PDB Molecule: adenylosuccinate lyase;PDBTitle: crystal structure of adenylosuccinate lyase (purb) from bacillus2 anthracis
43 c2x7aB_
not modelled
8.0
27
PDB header: immune systemChain: B: PDB Molecule: bone marrow stromal antigen 2;PDBTitle: structural basis of hiv-1 tethering to membranes by the2 bst2-tetherin ectodomain
44 d1v9va1
not modelled
8.0
11
Fold: Bromodomain-likeSuperfamily: MAST3 pre-PK domain-likeFamily: MAST3 pre-PK domain-like45 d2oeza1
not modelled
8.0
20
Fold: YacF-likeSuperfamily: YacF-likeFamily: YacF-like46 d1zk8a2
not modelled
7.9
5
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain47 c3rkoF_
not modelled
7.9
16
PDB header: oxidoreductaseChain: F: PDB Molecule: nadh-quinone oxidoreductase subunit j;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
48 c2j9wB_
not modelled
7.9
22
PDB header: protein transportChain: B: PDB Molecule: vps28-prov protein;PDBTitle: structural insight into the escrt-i-ii link and its role in2 mvb trafficking
49 c1y66D_
not modelled
7.7
27
PDB header: de novo proteinChain: D: PDB Molecule: engrailed homeodomain;PDBTitle: dioxane contributes to the altered conformation and2 oligomerization state of a designed engrailed homeodomain3 variant
50 d1c3ca_
not modelled
7.6
13
Fold: L-aspartase-likeSuperfamily: L-aspartase-likeFamily: L-aspartase/fumarase51 d2j9ua1
not modelled
7.5
22
Fold: Four-helical up-and-down bundleSuperfamily: VPS28 C-terminal domain-likeFamily: VPS28 C-terminal domain-like52 d2fcwa1
not modelled
7.4
18
Fold: RAP domain-likeSuperfamily: RAP domain-likeFamily: RAP domain53 c2qc7A_
not modelled
7.4
24
PDB header: chaperoneChain: A: PDB Molecule: endoplasmic reticulum protein erp29;PDBTitle: crystal structure of the protein-disulfide isomerase related chaperone2 erp29
54 c1q90R_
not modelled
7.2
16
PDB header: photosynthesisChain: R: PDB Molecule: cytochrome b6-f complex iron-sulfur subunit;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
55 d1q90r_
not modelled
7.2
16
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor56 c2qdqA_
not modelled
7.2
18
PDB header: structural proteinChain: A: PDB Molecule: talin-1;PDBTitle: crystal structure of the talin dimerisation domain
57 c2yvxD_
not modelled
7.1
8
PDB header: transport proteinChain: D: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte
58 c3no7A_
not modelled
7.1
28
PDB header: dna binding proteinChain: A: PDB Molecule: putative plasmid related protein;PDBTitle: crystal structure of the centromere-binding protein parb from plasmid2 pcxc100
59 d1i0aa_
not modelled
7.0
15
Fold: L-aspartase-likeSuperfamily: L-aspartase-likeFamily: L-aspartase/fumarase60 c1e3hA_
not modelled
6.9
12
PDB header: polyribonucleotide transferaseChain: A: PDB Molecule: guanosine pentaphosphate synthetase;PDBTitle: semet derivative of streptomyces antibioticus pnpase/gpsi2 enzyme
61 c1oy8A_
not modelled
6.8
13
PDB header: membrane proteinChain: A: PDB Molecule: acriflavine resistance protein b;PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
62 d1kx5c_
not modelled
6.8
27
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones63 d1iwga8
not modelled
6.7
7
Fold: Multidrug efflux transporter AcrB transmembrane domainSuperfamily: Multidrug efflux transporter AcrB transmembrane domainFamily: Multidrug efflux transporter AcrB transmembrane domain64 d1nh2d1
not modelled
6.7
18
Fold: Transcription factor IIA (TFIIA), alpha-helical domainSuperfamily: Transcription factor IIA (TFIIA), alpha-helical domainFamily: Transcription factor IIA (TFIIA), alpha-helical domain65 d2p12a1
not modelled
6.5
12
Fold: FomD barrel-likeSuperfamily: FomD-likeFamily: FomD-like66 d2oi8a2
not modelled
6.5
11
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain67 c3qh9A_
not modelled
6.4
16
PDB header: structural proteinChain: A: PDB Molecule: liprin-beta-2;PDBTitle: human liprin-beta2 coiled-coil
68 c2e9fC_
not modelled
6.3
24
PDB header: lyaseChain: C: PDB Molecule: argininosuccinate lyase;PDBTitle: crystal structure of t.th.hb8 argininosuccinate lyase complexed with2 l-arginine
69 c3zs9C_
not modelled
6.3
21
PDB header: hydrolase/transport proteinChain: C: PDB Molecule: golgi to er traffic protein 2;PDBTitle: s. cerevisiae get3-adp-alf4- complex with a cytosolic get2 fragment
70 d1q90g_
not modelled
6.3
18
Fold: Single transmembrane helixSuperfamily: PetG subunit of the cytochrome b6f complexFamily: PetG subunit of the cytochrome b6f complex71 c1q90G_
not modelled
6.3
18
PDB header: photosynthesisChain: G: PDB Molecule: cytochrome b6f complex subunit petg;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
72 c3sjdE_
not modelled
6.3
21
PDB header: hydrolase/transport proteinChain: E: PDB Molecule: golgi to er traffic protein 2;PDBTitle: crystal structure of s. cerevisiae get3 with bound adp-mg2+ in complex2 with get2 cytosolic domain
73 c3c8tA_
not modelled
6.3
11
PDB header: lyaseChain: A: PDB Molecule: fumarate lyase;PDBTitle: crystal structure of fumarate lyase from mesorhizobium sp. bnc1
74 d1id3c_
not modelled
6.3
27
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones75 d1eqza_
not modelled
6.2
27
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones76 c2p90B_
not modelled
6.2
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein cgl1923;PDBTitle: the crystal structure of a protein of unknown function from2 corynebacterium glutamicum atcc 13032
77 c2w6aB_
not modelled
6.2
16
PDB header: signaling proteinChain: B: PDB Molecule: arf gtpase-activating protein git1;PDBTitle: x-ray structure of the dimeric git1 coiled-coil domain
78 d2ftua1
not modelled
6.1
18
Fold: RAP domain-likeSuperfamily: RAP domain-likeFamily: RAP domain79 c3q5dA_
not modelled
6.1
12
PDB header: hydrolaseChain: A: PDB Molecule: atlastin-1;PDBTitle: crystal structure of human atlastin-1 (residues 1-447) bound to gdp,2 crystal form 1
80 d1nvpd1
not modelled
6.0
15
Fold: Transcription factor IIA (TFIIA), alpha-helical domainSuperfamily: Transcription factor IIA (TFIIA), alpha-helical domainFamily: Transcription factor IIA (TFIIA), alpha-helical domain81 c1s5lL_
not modelled
6.0
13
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
82 c3a0bl_
not modelled
6.0
13
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
83 c3kziL_
not modelled
6.0
13
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
84 c3prqL_
not modelled
6.0
13
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
85 c3prrL_
not modelled
6.0
13
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
86 c3a0hl_
not modelled
6.0
13
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
87 c3arcL_
not modelled
6.0
13
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
88 c3a0hL_
not modelled
6.0
13
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
89 c3a0bL_
not modelled
6.0
13
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
90 c1s5ll_
not modelled
6.0
13
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
91 c2axtL_
not modelled
6.0
13
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
92 c2axtl_
not modelled
6.0
13
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
93 c3bz2L_
not modelled
6.0
13
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 22 of 2). this file contains second monomer of psii dimer
94 c3bz1L_
not modelled
6.0
13
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
95 d2axtl1
not modelled
6.0
13
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein L, PsbLFamily: PsbL-like96 c3epvB_
not modelled
6.0
13
PDB header: metal binding proteinChain: B: PDB Molecule: nickel and cobalt resistance protein cnrr;PDBTitle: x-ray structure of the metal-sensor cnrx in both the apo- and copper-2 bound forms
97 d1q5na_
not modelled
5.9
11
Fold: L-aspartase-likeSuperfamily: L-aspartase-likeFamily: L-aspartase/fumarase98 d1tj7a_
not modelled
5.9
15
Fold: L-aspartase-likeSuperfamily: L-aspartase-likeFamily: L-aspartase/fumarase99 c3gaaB_
not modelled
5.9
2
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein ta1441;PDBTitle: the crystal structure of the protein with unknown function from2 thermoplasma acidophilum