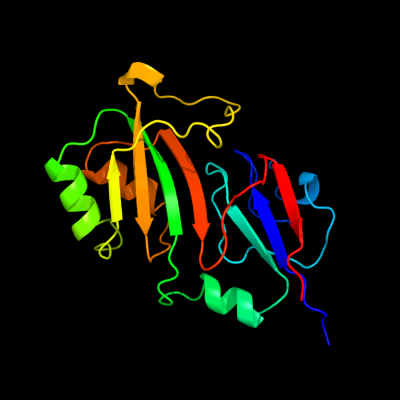
| 1 | c2omlA_
|
|
 |
100.0 |
94 |
PDB header:isomerase
Chain: A: PDB Molecule:ribosomal large subunit pseudouridine synthase e;
PDBTitle: crystal structure of e. coli pseudouridine synthase rlue
|
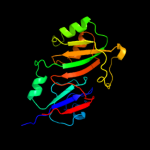
| 2 | c2olwB_
|
|
 |
100.0 |
95 |
PDB header:isomerase
Chain: B: PDB Molecule:ribosomal large subunit pseudouridine synthase e;
PDBTitle: crystal structure of e. coli pseudouridine synthase rlue
|
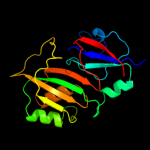
| 3 | c3dh3C_
|
|
 |
100.0 |
31 |
PDB header:isomerase/rna
Chain: C: PDB Molecule:ribosomal large subunit pseudouridine synthase f;
PDBTitle: crystal structure of rluf in complex with a 22 nucleotide2 rna substrate
|
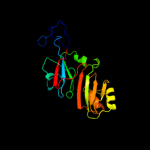
| 4 | c1vioA_
|
|
 |
100.0 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:ribosomal small subunit pseudouridine synthase a;
PDBTitle: crystal structure of pseudouridylate synthase
|
| 5 | c1kskA_
|
|
 |
100.0 |
32 |
PDB header:lyase
Chain: A: PDB Molecule:ribosomal small subunit pseudouridine synthase a;
PDBTitle: structure of rsua
|
| 6 | c2gmlA_
|
|
 |
100.0 |
35 |
PDB header:isomerase
Chain: A: PDB Molecule:ribosomal large subunit pseudouridine synthase f;
PDBTitle: crystal structure of catalytic domain of e.coli rluf
|
| 7 | d1vioa1
|
|
 |
100.0 |
29 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase RsuA/RluD |
| 8 | d1kska4
|
|
 |
100.0 |
36 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase RsuA/RluD |
| 9 | d1v9ka_
|
|
 |
100.0 |
17 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase RsuA/RluD |
| 10 | c1v9fA_
|
|
 |
100.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:ribosomal large subunit pseudouridine synthase d;
PDBTitle: crystal structure of catalytic domain of pseudouridine2 synthase rlud from escherichia coli
|
| 11 | d1v9fa_
|
|
 |
100.0 |
21 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase RsuA/RluD |
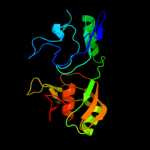
| 12 | c1qyuA_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:ribosomal large subunit pseudouridine synthase d;
PDBTitle: structure of the catalytic domain of 23s rrna pseudouridine2 synthase rlud
|
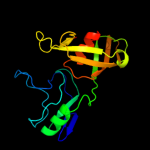
| 13 | c2i82D_
|
|
 |
100.0 |
20 |
PDB header:lyase/rna
Chain: D: PDB Molecule:ribosomal large subunit pseudouridine synthase a;
PDBTitle: crystal structure of pseudouridine synthase rlua: indirect2 sequence readout through protein-induced rna structure
|
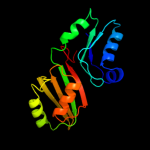
| 14 | d2apoa2
|
|
 |
98.8 |
13 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
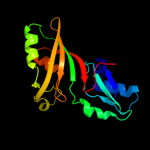
| 15 | d2ey4a2
|
|
 |
98.8 |
17 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
| 16 | c2ey4A_
|
|
 |
98.7 |
15 |
PDB header:isomerase/biosynthetic protein
Chain: A: PDB Molecule:probable trna pseudouridine synthase b;
PDBTitle: crystal structure of a cbf5-nop10-gar1 complex
|
| 17 | d1r3ea2
|
|
 |
98.6 |
18 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
| 18 | c2apoA_
|
|
 |
98.6 |
14 |
PDB header:isomerase/rna binding protein
Chain: A: PDB Molecule:probable trna pseudouridine synthase b;
PDBTitle: crystal structure of the methanococcus jannaschii cbf52 nop10 complex
|
| 19 | c3uaiA_
|
|
 |
98.5 |
15 |
PDB header:isomerase/chaperone
Chain: A: PDB Molecule:h/aca ribonucleoprotein complex subunit 4;
PDBTitle: structure of the shq1-cbf5-nop10-gar1 complex from saccharomyces2 cerevisiae
|
| 20 | d1sgva2
|
|
 |
98.5 |
18 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
| 21 | d1k8wa5 |
|
not modelled |
98.5 |
20 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
| 22 | c1sgvA_ |
|
not modelled |
98.4 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:trna pseudouridine synthase b;
PDBTitle: structure of trna psi55 pseudouridine synthase (trub)
|
| 23 | c1k8wA_ |
|
not modelled |
98.0 |
21 |
PDB header:lyase/rna
Chain: A: PDB Molecule:trna pseudouridine synthase b;
PDBTitle: crystal structure of the e. coli pseudouridine synthase2 trub bound to a t stem-loop rna
|
| 24 | c1ze2B_ |
|
not modelled |
95.4 |
18 |
PDB header:lyase/rna
Chain: B: PDB Molecule:trna pseudouridine synthase b;
PDBTitle: conformational change of pseudouridine 55 synthase upon its2 association with rna substrate
|
| 25 | c2k6pA_ |
|
not modelled |
32.2 |
11 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein hp_1423;
PDBTitle: solution structure of hypothetical protein, hp1423
|
| 26 | c2fo1A_ |
|
not modelled |
19.2 |
16 |
PDB header:gene regulation/signalling protein/dna
Chain: A: PDB Molecule:lin-12 and glp-1 phenotype protein 1, isoform b;
PDBTitle: crystal structure of the csl-notch-mastermind ternary2 complex bound to dna
|
| 27 | d2ysca1 |
|
not modelled |
13.8 |
45 |
Fold:WW domain-like
Superfamily:WW domain
Family:WW domain |
| 28 | d1nkga1 |
|
not modelled |
12.4 |
14 |
Fold:Prealbumin-like
Superfamily:Starch-binding domain-like
Family:Rhamnogalacturonase B, RhgB, middle domain |
| 29 | c3cjsA_ |
|
not modelled |
12.0 |
50 |
PDB header:transferase/ribosomal protein
Chain: A: PDB Molecule:ribosomal protein l11 methyltransferase;
PDBTitle: minimal recognition complex between prma and ribosomal protein l11
|
| 30 | d2ho2a1 |
|
not modelled |
11.7 |
42 |
Fold:WW domain-like
Superfamily:WW domain
Family:WW domain |
| 31 | c2jmzA_ |
|
not modelled |
11.6 |
14 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein mj0781;
PDBTitle: solution structure of a klba intein precursor from2 methanococcus jannaschii
|
| 32 | c1f5nA_ |
|
not modelled |
11.4 |
13 |
PDB header:signaling protein
Chain: A: PDB Molecule:interferon-induced guanylate-binding protein 1;
PDBTitle: human guanylate binding protein-1 in complex with the gtp2 analogue, gmppnp.
|
| 33 | c3h7hA_ |
|
not modelled |
11.2 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor spt4;
PDBTitle: crystal structure of the human transcription elongation factor dsif,2 hspt4/hspt5 (176-273)
|
| 34 | d1zhva1 |
|
not modelled |
9.4 |
31 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Atu0741-like |
| 35 | c1mzwB_ |
|
not modelled |
8.3 |
23 |
PDB header:isomerase
Chain: B: PDB Molecule:u4/u6 snrnp 60kda protein;
PDBTitle: crystal structure of a u4/u6 snrnp complex between human2 spliceosomal cyclophilin h and a u4/u6-60k peptide
|
| 36 | d2hq4a1 |
|
not modelled |
7.9 |
33 |
Fold:PH1570-like
Superfamily:PH1570-like
Family:PH1570-like |
| 37 | c2voiB_ |
|
not modelled |
7.8 |
27 |
PDB header:apoptosis
Chain: B: PDB Molecule:bh3-interacting domain death agonist p13;
PDBTitle: structure of mouse a1 bound to the bid bh3-domain
|
| 38 | c2imzA_ |
|
not modelled |
6.6 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease pi-mtui;
PDBTitle: crystal structure of mtu reca intein splicing domain
|
| 39 | d1t6la1 |
|
not modelled |
6.4 |
19 |
Fold:DNA clamp
Superfamily:DNA clamp
Family:DNA polymerase processivity factor |
| 40 | c2exuA_ |
|
not modelled |
6.2 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:transcription initiation protein spt4/spt5;
PDBTitle: crystal structure of saccharomyces cerevisiae transcription elongation2 factors spt4-spt5ngn domain
|
| 41 | c1tlqA_ |
|
not modelled |
6.1 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ypjq;
PDBTitle: crystal structure of protein ypjq from bacillus subtilis, pfam duf64
|
| 42 | d1tlqa_ |
|
not modelled |
6.1 |
13 |
Fold:YutG-like
Superfamily:YutG-like
Family:YutG-like |
| 43 | d1k4ta3 |
|
not modelled |
5.4 |
13 |
Fold:Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragment
Superfamily:Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragment
Family:Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragment |
| 44 | d1y9ia_ |
|
not modelled |
5.3 |
13 |
Fold:YutG-like
Superfamily:YutG-like
Family:YutG-like |
| 45 | c3bbnD_ |
|
not modelled |
5.3 |
13 |
PDB header:ribosome
Chain: D: PDB Molecule:ribosomal protein s4;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|
| 46 | c1vs3B_ |
|
not modelled |
5.2 |
40 |
PDB header:isomerase
Chain: B: PDB Molecule:trna pseudouridine synthase a;
PDBTitle: crystal structure of the trna pseudouridine synthase trua from thermus2 thermophilus hb8
|
| 47 | c2x5pA_ |
|
not modelled |
5.2 |
18 |
PDB header:protein binding
Chain: A: PDB Molecule:fibronectin binding protein;
PDBTitle: crystal structure of the streptococcus pyogenes fibronectin binding2 protein fbab-b
|