| 1 | c2pptA_ |
|
 |
94.6 |
17 |
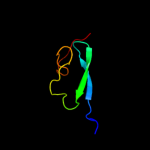
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin-2;
PDBTitle: crystal structure of thioredoxin-2
|
| 2 | c2lcqA_ |
|
 |
92.8 |
25 |
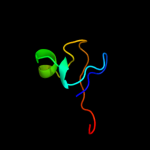
PDB header:metal binding protein
Chain: A: PDB Molecule:putative toxin vapc6;
PDBTitle: solution structure of the endonuclease nob1 from p.horikoshii
|
| 3 | c2hr5B_ |
|
 |
92.0 |
30 |
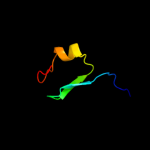
PDB header:metal binding protein
Chain: B: PDB Molecule:rubrerythrin;
PDBTitle: pf1283- rubrerythrin from pyrococcus furiosus iron bound form
|
| 4 | d1dl6a_ |
|
 |
91.1 |
18 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Transcriptional factor domain |
| 5 | c2f9iD_ |
|
 |
90.8 |
17 |
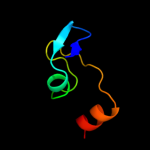
PDB header:transferase
Chain: D: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl
PDBTitle: crystal structure of the carboxyltransferase subunit of acc2 from staphylococcus aureus
|
| 6 | d1akya2 |
|
 |
90.5 |
5 |
Fold:Rubredoxin-like
Superfamily:Microbial and mitochondrial ADK, insert "zinc finger" domain
Family:Microbial and mitochondrial ADK, insert "zinc finger" domain |
| 7 | d2ak3a2 |
|
 |
89.8 |
6 |
Fold:Rubredoxin-like
Superfamily:Microbial and mitochondrial ADK, insert "zinc finger" domain
Family:Microbial and mitochondrial ADK, insert "zinc finger" domain |
| 8 | d1p3ja2 |
|
 |
89.4 |
12 |
Fold:Rubredoxin-like
Superfamily:Microbial and mitochondrial ADK, insert "zinc finger" domain
Family:Microbial and mitochondrial ADK, insert "zinc finger" domain |
| 9 | d2f9yb1 |
|
 |
89.1 |
22 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 10 | c2f9yB_ |
|
 |
89.1 |
22 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl transferase subunit
PDBTitle: the crystal structure of the carboxyltransferase subunit of acc from2 escherichia coli
|
| 11 | d1zina2 |
|
 |
88.9 |
15 |
Fold:Rubredoxin-like
Superfamily:Microbial and mitochondrial ADK, insert "zinc finger" domain
Family:Microbial and mitochondrial ADK, insert "zinc finger" domain |
| 12 | d1s3ga2 |
|
 |
88.8 |
12 |
Fold:Rubredoxin-like
Superfamily:Microbial and mitochondrial ADK, insert "zinc finger" domain
Family:Microbial and mitochondrial ADK, insert "zinc finger" domain |
| 13 | c1dvbA_ |
|
 |
87.6 |
22 |
PDB header:electron transport
Chain: A: PDB Molecule:rubrerythrin;
PDBTitle: rubrerythrin
|
| 14 | d1k3xa3 |
|
 |
87.4 |
27 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:C-terminal, Zn-finger domain of MutM-like DNA repair proteins |
| 15 | d1yc5a1 |
|
 |
87.3 |
24 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 16 | d1e4va2 |
|
 |
86.7 |
0 |
Fold:Rubredoxin-like
Superfamily:Microbial and mitochondrial ADK, insert "zinc finger" domain
Family:Microbial and mitochondrial ADK, insert "zinc finger" domain |
| 17 | c1hk8A_ |
|
 |
86.4 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:anaerobic ribonucleotide-triphosphate reductase;
PDBTitle: structural basis for allosteric substrate specificity2 regulation in class iii ribonucleotide reductases:3 nrdd in complex with dgtp
|
| 18 | d1hk8a_ |
|
 |
86.4 |
14 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:Class III anaerobic ribonucleotide reductase NRDD subunit |
| 19 | c2gb5B_ |
|
 |
86.0 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:nadh pyrophosphatase;
PDBTitle: crystal structure of nadh pyrophosphatase (ec 3.6.1.22) (1790429) from2 escherichia coli k12 at 2.30 a resolution
|
| 20 | d2gnra1 |
|
 |
86.0 |
29 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:SSO2064-like |
| 21 | d1ee8a3 |
|
not modelled |
85.5 |
26 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:C-terminal, Zn-finger domain of MutM-like DNA repair proteins |
| 22 | c3na7A_ |
|
not modelled |
85.4 |
18 |
PDB header:gene regulation, chaperone
Chain: A: PDB Molecule:hp0958;
PDBTitle: 2.2 angstrom structure of the hp0958 protein from helicobacter pylori2 ccug 17874
|
| 23 | d1pfta_ |
|
not modelled |
84.9 |
17 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Transcriptional factor domain |
| 24 | d1m2ka_ |
|
not modelled |
84.4 |
21 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 25 | d2gmga1 |
|
not modelled |
84.0 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:PF0610-like |
| 26 | c3glsC_ |
|
not modelled |
83.7 |
21 |
PDB header:hydrolase
Chain: C: PDB Molecule:nad-dependent deacetylase sirtuin-3,
PDBTitle: crystal structure of human sirt3
|
| 27 | c2kdxA_ |
|
not modelled |
83.5 |
29 |
PDB header:metal-binding protein
Chain: A: PDB Molecule:hydrogenase/urease nickel incorporation protein
PDBTitle: solution structure of hypa protein
|
| 28 | c3a44D_ |
|
not modelled |
83.4 |
30 |
PDB header:metal binding protein
Chain: D: PDB Molecule:hydrogenase nickel incorporation protein hypa;
PDBTitle: crystal structure of hypa in the dimeric form
|
| 29 | d1tdza3 |
|
not modelled |
83.2 |
26 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:C-terminal, Zn-finger domain of MutM-like DNA repair proteins |
| 30 | c3lpeF_ |
|
not modelled |
82.1 |
24 |
PDB header:transferase
Chain: F: PDB Molecule:dna-directed rna polymerase subunit e'';
PDBTitle: crystal structure of spt4/5ngn heterodimer complex from methanococcus2 jannaschii
|
| 31 | c3k7aM_ |
|
not modelled |
81.9 |
24 |
PDB header:transcription
Chain: M: PDB Molecule:transcription initiation factor iib;
PDBTitle: crystal structure of an rna polymerase ii-tfiib complex
|
| 32 | d1lkoa2 |
|
not modelled |
81.5 |
25 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 33 | c3k35D_ |
|
not modelled |
81.4 |
19 |
PDB header:hydrolase
Chain: D: PDB Molecule:nad-dependent deacetylase sirtuin-6;
PDBTitle: crystal structure of human sirt6
|
| 34 | c3p2aB_ |
|
not modelled |
81.3 |
32 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative thioredoxin-like protein;
PDBTitle: crystal structure of thioredoxin 2 from yersinia pestis
|
| 35 | d2ct7a1 |
|
not modelled |
80.6 |
28 |
Fold:RING/U-box
Superfamily:RING/U-box
Family:IBR domain |
| 36 | d1k82a3 |
|
not modelled |
80.5 |
24 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:C-terminal, Zn-finger domain of MutM-like DNA repair proteins |
| 37 | d1l1ta3 |
|
not modelled |
80.5 |
32 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:C-terminal, Zn-finger domain of MutM-like DNA repair proteins |
| 38 | d1q1aa_ |
|
not modelled |
80.4 |
21 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 39 | d1r2za3 |
|
not modelled |
79.2 |
32 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:C-terminal, Zn-finger domain of MutM-like DNA repair proteins |
| 40 | d1weoa_ |
|
not modelled |
78.7 |
22 |
Fold:RING/U-box
Superfamily:RING/U-box
Family:RING finger domain, C3HC4 |
| 41 | c2ja6L_ |
|
not modelled |
77.6 |
26 |
PDB header:transferase
Chain: L: PDB Molecule:dna-directed rna polymerases i, ii, and iii 7.7
PDBTitle: cpd lesion containing rna polymerase ii elongation complex2 b
|
| 42 | c3h0gL_ |
|
not modelled |
77.5 |
19 |
PDB header:transcription
Chain: L: PDB Molecule:dna-directed rna polymerases i, ii, and iii
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
|
| 43 | d2akla2 |
|
not modelled |
76.3 |
28 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:PhnA zinc-binding domain |
| 44 | c2aklA_ |
|
not modelled |
76.0 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phna-like protein pa0128;
PDBTitle: solution structure for phn-a like protein pa0128 from2 pseudomonas aeruginosa
|
| 45 | c3pkiF_ |
|
not modelled |
75.9 |
16 |
PDB header:hydrolase
Chain: F: PDB Molecule:nad-dependent deacetylase sirtuin-6;
PDBTitle: human sirt6 crystal structure in complex with adp ribose
|
| 46 | c1i3qI_ |
|
not modelled |
75.2 |
19 |
PDB header:transcription
Chain: I: PDB Molecule:dna-directed rna polymerase ii 14.2kd
PDBTitle: rna polymerase ii crystal form i at 3.1 a resolution
|
| 47 | c3axtA_ |
|
not modelled |
75.1 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:probable n(2),n(2)-dimethylguanosine trna methyltransferase
PDBTitle: complex structure of trna methyltransferase trm1 from aquifex aeolicus2 with s-adenosyl-l-methionine
|
| 48 | d1yuza2 |
|
not modelled |
74.5 |
22 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 49 | c3p2aA_ |
|
not modelled |
73.1 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative thioredoxin-like protein;
PDBTitle: crystal structure of thioredoxin 2 from yersinia pestis
|
| 50 | d6rxna_ |
|
not modelled |
72.9 |
23 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 51 | c2jrpA_ |
|
not modelled |
72.2 |
37 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative cytoplasmic protein;
PDBTitle: solution nmr structure of yfgj from salmonella typhimurium2 modeled with two zn+2 bound, northeast structural genomics3 consortium target str86
|
| 52 | d2dsxa1 |
|
not modelled |
72.2 |
26 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 53 | c3cngC_ |
|
not modelled |
72.0 |
25 |
PDB header:hydrolase
Chain: C: PDB Molecule:nudix hydrolase;
PDBTitle: crystal structure of nudix hydrolase from nitrosomonas europaea
|
| 54 | d1qcva_ |
|
not modelled |
71.7 |
11 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 55 | c1q14A_ |
|
not modelled |
71.7 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:hst2 protein;
PDBTitle: structure and autoregulation of the yeast hst2 homolog of sir2
|
| 56 | d2dkta1 |
|
not modelled |
71.2 |
24 |
Fold:CHY zinc finger-like
Superfamily:CHY zinc finger-like
Family:CHY zinc finger |
| 57 | d4rxna_ |
|
not modelled |
70.9 |
22 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 58 | c2x5cB_ |
|
not modelled |
70.5 |
60 |
PDB header:viral protein
Chain: B: PDB Molecule:hypothetical protein orf131;
PDBTitle: crystal structure of hypothetical protein orf131 from2 pyrobaculum spherical virus
|
| 59 | d1iroa_ |
|
not modelled |
70.0 |
22 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 60 | c3jwpA_ |
|
not modelled |
68.9 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulatory protein sir2 homologue;
PDBTitle: crystal structure of plasmodium falciparum sir2a (pf13_0152) in2 complex with amp
|
| 61 | d2k4xa1 |
|
not modelled |
68.6 |
25 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein S27a |
| 62 | c2v3bB_ |
|
not modelled |
68.3 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:rubredoxin 2;
PDBTitle: crystal structure of the electron transfer complex2 rubredoxin - rubredoxin reductase from pseudomonas3 aeruginosa.
|
| 63 | d2cona1 |
|
 |
68.0 |
28 |
Fold:Rubredoxin-like
Superfamily:NOB1 zinc finger-like
Family:NOB1 zinc finger-like |
| 64 | c1s24A_ |
|
not modelled |
68.0 |
30 |
PDB header:electron transport
Chain: A: PDB Molecule:rubredoxin 2;
PDBTitle: rubredoxin domain ii from pseudomonas oleovorans
|
| 65 | d1s24a_ |
|
not modelled |
68.0 |
30 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 66 | c2js4A_ |
|
not modelled |
67.8 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0434 protein bb2007;
PDBTitle: solution nmr structure of bordetella bronchiseptica protein2 bb2007. northeast structural genomics consortium target3 bor54
|
| 67 | c2odxA_ |
|
 |
67.8 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome c oxidase polypeptide iv;
PDBTitle: solution structure of zn(ii)cox4
|
| 68 | d2ey4e1 |
|
not modelled |
67.1 |
22 |
Fold:Rubredoxin-like
Superfamily:Nop10-like SnoRNP
Family:Nucleolar RNA-binding protein Nop10-like |
| 69 | d1p91a_ |
|
not modelled |
67.0 |
33 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:rRNA methyltransferase RlmA |
| 70 | d1nnqa2 |
|
not modelled |
66.8 |
35 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 71 | c1yuzB_ |
|
 |
66.8 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nigerythrin;
PDBTitle: partially reduced state of nigerythrin
|
| 72 | d1qxfa_ |
|
not modelled |
66.3 |
14 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein S27e |
| 73 | d2apob1 |
|
not modelled |
66.2 |
27 |
Fold:Rubredoxin-like
Superfamily:Nop10-like SnoRNP
Family:Nucleolar RNA-binding protein Nop10-like |
| 74 | d2pk7a1 |
|
not modelled |
66.0 |
19 |
Fold:Trm112p-like
Superfamily:Trm112p-like
Family:Trm112p-like |
| 75 | c2kn9A_ |
|
not modelled |
66.0 |
33 |
PDB header:electron transport
Chain: A: PDB Molecule:rubredoxin;
PDBTitle: solution structure of zinc-substituted rubredoxin b (rv3250c) from2 mycobacterium tuberculosis. seattle structural genomics center for3 infectious disease target mytud.01635.a
|
| 76 | c2jr6A_ |
|
not modelled |
65.9 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0434 protein nma0874;
PDBTitle: solution structure of upf0434 protein nma0874. northeast structural2 genomics target mr32
|
| 77 | c3zyqA_ |
|
 |
65.6 |
22 |
PDB header:signaling
Chain: A: PDB Molecule:hepatocyte growth factor-regulated tyrosine kinase
PDBTitle: crystal structure of the tandem vhs and fyve domains of hepatocyte2 growth factor-regulated tyrosine kinase substrate (hgs-hrs) at 1.483 a resolution
|
| 78 | c2pziA_ |
|
not modelled |
65.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:probable serine/threonine-protein kinase pkng;
PDBTitle: crystal structure of protein kinase pkng from mycobacterium2 tuberculosis in complex with tetrahydrobenzothiophene ax20017
|
| 79 | c3floD_ |
|
not modelled |
64.8 |
29 |
PDB header:transferase
Chain: D: PDB Molecule:dna polymerase alpha catalytic subunit a;
PDBTitle: crystal structure of the carboxyl-terminal domain of yeast2 dna polymerase alpha in complex with its b subunit
|
| 80 | d1ryqa_ |
|
 |
64.6 |
36 |
Fold:Rubredoxin-like
Superfamily:RNA polymerase subunits
Family:RpoE2-like |
| 81 | d2jnya1 |
|
not modelled |
64.5 |
19 |
Fold:Trm112p-like
Superfamily:Trm112p-like
Family:Trm112p-like |
| 82 | d2fiya1 |
|
not modelled |
64.3 |
22 |
Fold:FdhE-like
Superfamily:FdhE-like
Family:FdhE-like |
| 83 | c3ky9B_ |
|
 |
63.8 |
29 |
PDB header:apoptosis
Chain: B: PDB Molecule:proto-oncogene vav;
PDBTitle: autoinhibited vav1
|
| 84 | d1dx8a_ |
|
not modelled |
63.3 |
22 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 85 | d1h7va_ |
|
not modelled |
62.0 |
22 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 86 | c3gn5B_ |
|
not modelled |
61.3 |
25 |
PDB header:dna binding protein
Chain: B: PDB Molecule:hth-type transcriptional regulator mqsa (ygit/b3021);
PDBTitle: structure of the e. coli protein mqsa (ygit/b3021)
|
| 87 | d1iu5a_ |
|
not modelled |
61.0 |
26 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 88 | d2rdva_ |
|
not modelled |
58.8 |
26 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 89 | d2j0151 |
|
not modelled |
57.6 |
27 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein L32p |
| 90 | c3iz6X_ |
|
not modelled |
57.3 |
20 |
PDB header:ribosome
Chain: X: PDB Molecule:40s ribosomal protein s27 (s27e);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 91 | d2hf1a1 |
|
not modelled |
57.0 |
19 |
Fold:Trm112p-like
Superfamily:Trm112p-like
Family:Trm112p-like |
| 92 | c3f2cA_ |
|
not modelled |
56.6 |
22 |
PDB header:transferase/dna
Chain: A: PDB Molecule:geobacillus kaustophilus dna polc;
PDBTitle: dna polymerase polc from geobacillus kaustophilus complex with dna,2 dgtp and mn
|
| 93 | d1rb9a_ |
|
not modelled |
56.3 |
30 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 94 | c3h0gI_ |
|
not modelled |
55.0 |
15 |
PDB header:transcription
Chain: I: PDB Molecule:dna-directed rna polymerase ii subunit rpb9;
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
|
| 95 | d1brfa_ |
|
not modelled |
54.9 |
32 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 96 | d2zjrz1 |
|
not modelled |
54.4 |
33 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein L32p |
| 97 | d1j8fa_ |
|
not modelled |
53.8 |
22 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 98 | c2opfA_ |
|
not modelled |
53.7 |
27 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:endonuclease viii;
PDBTitle: crystal structure of the dna repair enzyme endonuclease-viii (nei)2 from e. coli (r252a) in complex with ap-site containing dna substrate
|
| 99 | d1ibia1 |
|
not modelled |
53.6 |
30 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 100 | c1k82D_ |
|
not modelled |
53.4 |
20 |
PDB header:hydrolase/dna
Chain: D: PDB Molecule:formamidopyrimidine-dna glycosylase;
PDBTitle: crystal structure of e.coli formamidopyrimidine-dna2 glycosylase (fpg) covalently trapped with dna
|
| 101 | d2ayja1 |
|
not modelled |
53.3 |
24 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein L40e |
| 102 | d1l1oc_ |
|
not modelled |
52.0 |
24 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 103 | c2kpiA_ |
|
not modelled |
51.6 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein sco3027;
PDBTitle: solution nmr structure of streptomyces coelicolor sco30272 modeled with zn+2 bound, northeast structural genomics3 consortium target rr58
|
| 104 | c1nnjA_ |
|
not modelled |
51.3 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:formamidopyrimidine-dna glycosylase;
PDBTitle: crystal structure complex between the lactococcus lactis fpg and an2 abasic site containing dna
|
| 105 | d2jnea1 |
|
not modelled |
50.7 |
24 |
Fold:Rubredoxin-like
Superfamily:YfgJ-like
Family:YfgJ-like |
| 106 | c2jneA_ |
|
not modelled |
50.7 |
24 |
PDB header:metal binding protein
Chain: A: PDB Molecule:hypothetical protein yfgj;
PDBTitle: nmr structure of e.coli yfgj modelled with two zn+2 bound.2 northeast structural genomics consortium target er317.
|
| 107 | d1x6ma_ |
|
not modelled |
50.4 |
15 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:Glutathione-dependent formaldehyde-activating enzyme, Gfa |
| 108 | c3ir9A_ |
|
not modelled |
50.0 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:peptide chain release factor subunit 1;
PDBTitle: c-terminal domain of peptide chain release factor from2 methanosarcina mazei.
|
| 109 | c3nw0A_ |
|
not modelled |
48.7 |
25 |
PDB header:metal binding protein
Chain: A: PDB Molecule:non-structural maintenance of chromosomes element 1
PDBTitle: crystal structure of mageg1 and nse1 complex
|
| 110 | d1ma3a_ |
|
not modelled |
47.4 |
18 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 111 | c3u50C_ |
|
not modelled |
46.7 |
28 |
PDB header:dna binding protein
Chain: C: PDB Molecule:telomerase-associated protein 82;
PDBTitle: crystal structure of the tetrahymena telomerase processivity factor2 teb1 ob-c
|
| 112 | d2qam01 |
|
not modelled |
46.5 |
11 |
Fold:Rubredoxin-like
Superfamily:Zn-binding ribosomal proteins
Family:Ribosomal protein L32p |
| 113 | c3h7hA_ |
|
not modelled |
46.5 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor spt4;
PDBTitle: crystal structure of the human transcription elongation factor dsif,2 hspt4/hspt5 (176-273)
|
| 114 | c2f5qA_ |
|
not modelled |
46.3 |
27 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:formamidopyrimidine-dna glycosidase;
PDBTitle: catalytically inactive (e3q) mutm crosslinked to oxog:c2 containing dna cc2
|
| 115 | d1pfva3 |
|
not modelled |
45.9 |
29 |
Fold:Rubredoxin-like
Superfamily:Methionyl-tRNA synthetase (MetRS), Zn-domain
Family:Methionyl-tRNA synthetase (MetRS), Zn-domain |
| 116 | d1ctla1 |
|
not modelled |
45.8 |
36 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 117 | c1ee8A_ |
|
not modelled |
45.8 |
20 |
PDB header:dna binding protein
Chain: A: PDB Molecule:mutm (fpg) protein;
PDBTitle: crystal structure of mutm (fpg) protein from thermus thermophilus hb8
|
| 118 | d1dgsa1 |
|
not modelled |
45.4 |
27 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:NAD+-dependent DNA ligase, domain 3 |
| 119 | c3izbX_ |
|
not modelled |
45.2 |
14 |
PDB header:ribosome
Chain: X: PDB Molecule:40s ribosomal protein rps27 (s27e);
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 120 | c1ltlE_ |
|
not modelled |
43.5 |
14 |
PDB header:replication
Chain: E: PDB Molecule:dna replication initiator (cdc21/cdc54);
PDBTitle: the dodecamer structure of mcm from archaeal m.2 thermoautotrophicum
|

