1 c1o98A_
100.0
50
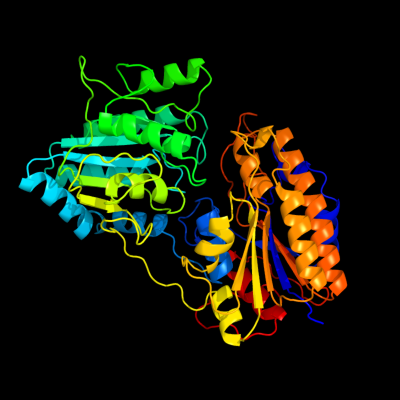
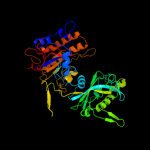
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independentPDBTitle: 1.4a crystal structure of phosphoglycerate mutase from2 bacillus stearothermophilus complexed with3 2-phosphoglycerate
2 c3igzB_
100.0
34
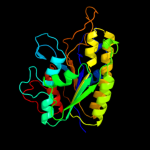
PDB header: isomeraseChain: B: PDB Molecule: cofactor-independent phosphoglycerate mutase;PDBTitle: crystal structures of leishmania mexicana phosphoglycerate2 mutase at low cobalt concentration
3 d1o98a1
100.0
42
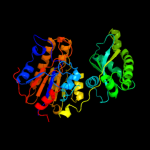
Fold: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domainSuperfamily: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domainFamily: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain4 c3iddA_
100.0
20
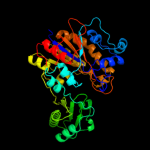
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independentPDBTitle: cofactor-independent phosphoglycerate mutase from2 thermoplasma acidophilum dsm 1728
5 d1o98a2
100.0
60
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, catalytic domain6 c2zktB_
100.0
21
PDB header: isomeraseChain: B: PDB Molecule: 2,3-bisphosphoglycerate-independent phosphoglyceratePDBTitle: structure of ph0037 protein from pyrococcus horikoshii
7 c2i09A_
100.0
18
PDB header: isomeraseChain: A: PDB Molecule: phosphopentomutase;PDBTitle: crystal structure of putative phosphopentomutase from streptococcus2 mutans
8 c3m8yC_
99.9
25
PDB header: isomeraseChain: C: PDB Molecule: phosphopentomutase;PDBTitle: phosphopentomutase from bacillus cereus after glucose-1,6-bisphosphate2 activation
9 d2i09a1
99.9
26
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: DeoB catalytic domain-like10 c3lxqB_
99.8
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein vp1736;PDBTitle: the crystal structure of a protein in the alkaline2 phosphatase superfamily from vibrio parahaemolyticus to3 1.95a
11 d1fsua_
99.8
13
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase12 c3q3qA_
99.8
29
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase;PDBTitle: crystal structure of spap: an novel alkaline phosphatase from2 bacterium sphingomonas sp. strain bsar-1
13 d1hdha_
99.8
21
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase14 c3b5qB_
99.7
21
PDB header: hydrolaseChain: B: PDB Molecule: putative sulfatase yidj;PDBTitle: crystal structure of a putative sulfatase (np_810509.1)2 from bacteroides thetaiotaomicron vpi-5482 at 2.40 a3 resolution
15 d1auka_
99.7
16
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase16 c2qzuA_
99.7
13
PDB header: hydrolaseChain: A: PDB Molecule: putative sulfatase yidj;PDBTitle: crystal structure of the putative sulfatase yidj from bacteroides2 fragilis. northeast structural genomics consortium target bfr123
17 c3ed4A_
99.7
17
PDB header: transferaseChain: A: PDB Molecule: arylsulfatase;PDBTitle: crystal structure of putative arylsulfatase from escherichia coli
18 d1p49a_
99.7
19
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase19 c2vqrA_
99.7
20
PDB header: hydrolaseChain: A: PDB Molecule: putative sulfatase;PDBTitle: crystal structure of a phosphonate monoester hydrolase2 from rhizobium leguminosarum: a new member of the3 alkaline phosphatase superfamily
20 c2w5tA_
99.7
14
PDB header: transferaseChain: A: PDB Molecule: processed glycerol phosphate lipoteichoic acidPDBTitle: structure-based mechanism of lipoteichoic acid synthesis by2 staphylococcus aureus ltas.
21 c2xrgA_
not modelled
99.7
13
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: crystal structure of autotaxin (enpp2) in complex with the2 ha155 boronic acid inhibitor
22 c2w8dB_
not modelled
99.7
13
PDB header: transferaseChain: B: PDB Molecule: processed glycerol phosphate lipoteichoic acid synthase 2;PDBTitle: distinct and essential morphogenic functions for wall- and2 lipo-teichoic acids in bacillus subtilis
23 c2xr9A_
not modelled
99.7
13
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: crystal structure of autotaxin (enpp2)
24 d1y6va1
not modelled
99.5
20
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase25 c2iucB_
not modelled
99.4
12
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: structure of alkaline phosphatase from the antarctic2 bacterium tab5
26 c3a52A_
not modelled
99.4
26
PDB header: hydrolaseChain: A: PDB Molecule: cold-active alkaline phosphatase;PDBTitle: crystal structure of cold-active alkailne phosphatase from2 psychrophile shewanella sp.
27 c1ew2A_
not modelled
99.4
17
PDB header: hydrolaseChain: A: PDB Molecule: phosphatase;PDBTitle: crystal structure of a human phosphatase
28 d1k7ha_
not modelled
99.4
21
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase29 d1zeda1
not modelled
99.4
17
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase30 c2gsoB_
not modelled
99.3
25
PDB header: hydrolaseChain: B: PDB Molecule: phosphodiesterase-nucleotide pyrophosphatase;PDBTitle: structure of xac nucleotide2 pyrophosphatase/phosphodiesterase in complex with vanadate
31 c2w0yB_
not modelled
99.3
15
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: h.salinarum alkaline phosphatase
32 c2x98A_
not modelled
99.3
16
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase;PDBTitle: h.salinarum alkaline phosphatase
33 c3e2dB_
not modelled
99.0
19
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: the 1.4 a crystal structure of the large and cold-active2 vibrio sp. alkaline phosphatase
34 d1ei6a_
not modelled
98.2
16
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Phosphonoacetate hydrolase35 c3szzA_
not modelled
97.7
24
PDB header: hydrolaseChain: A: PDB Molecule: phosphonoacetate hydrolase;PDBTitle: crystal structure of phosphonoacetate hydrolase from sinorhizobium2 meliloti 1021 in complex with acetate
36 c2d1gB_
not modelled
97.4
9
PDB header: hydrolaseChain: B: PDB Molecule: acid phosphatase;PDBTitle: structure of francisella tularensis acid phosphatase a (acpa) bound to2 orthovanadate
37 c3m0zD_
not modelled
91.9
15
PDB header: lyaseChain: D: PDB Molecule: putative aldolase;PDBTitle: crystal structure of putative aldolase from klebsiella2 pneumoniae.
38 c3m6yA_
not modelled
79.6
18
PDB header: lyaseChain: A: PDB Molecule: 4-hydroxy-2-oxoglutarate aldolase;PDBTitle: structure of 4-hydroxy-2-oxoglutarate aldolase from bacillus cereus at2 1.45 a resolution.
39 c3muxB_
not modelled
75.3
18
PDB header: lyaseChain: B: PDB Molecule: putative 4-hydroxy-2-oxoglutarate aldolase;PDBTitle: the crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase2 from bacillus anthracis to 1.45a
40 c1qzwC_
not modelled
47.3
18
PDB header: signaling protein/rnaChain: C: PDB Molecule: signal recognition 54 kda protein;PDBTitle: crystal structure of the complete core of archaeal srp and2 implications for inter-domain communication
41 d1vyva2
not modelled
46.8
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases42 c3ib7A_
not modelled
46.1
16
PDB header: hydrolaseChain: A: PDB Molecule: icc protein;PDBTitle: crystal structure of full length rv0805
43 c3tovB_
not modelled
44.0
29
PDB header: transferaseChain: B: PDB Molecule: glycosyl transferase family 9;PDBTitle: the crystal structure of the glycosyl transferase family 9 from2 veillonella parvula dsm 2008
44 c2j37W_
not modelled
43.9
17
PDB header: ribosomeChain: W: PDB Molecule: signal recognition particle 54 kda proteinPDBTitle: model of mammalian srp bound to 80s rncs
45 c2iy3A_
not modelled
28.9
15
PDB header: rna-bindingChain: A: PDB Molecule: signal recognition particle protein ffh;PDBTitle: structure of the e. coli signal recognition particle2 bound to a translating ribosome
46 d2nxfa1
not modelled
27.8
16
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: ADPRibase-Mn-like47 d1vyua2
not modelled
26.9
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases48 d2z3va1
not modelled
24.3
22
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like49 d1iq0a1
not modelled
24.0
33
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases50 d1whua_
not modelled
23.8
55
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3Family: Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 351 d1o57a1
not modelled
23.8
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: N-terminal domain of Bacillus PurR52 d1f7ua1
not modelled
23.3
17
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases53 d1ydga_
not modelled
22.7
9
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like54 c2ip1A_
not modelled
22.1
16
PDB header: ligaseChain: A: PDB Molecule: tryptophanyl-trna synthetase;PDBTitle: crystal structure analysis of s. cerevisiae tryptophanyl trna2 synthetase
55 c2j289_
not modelled
21.2
15
PDB header: ribosomeChain: 9: PDB Molecule: signal recognition particle 54;PDBTitle: model of e. coli srp bound to 70s rncs
56 c3dm5A_
not modelled
20.0
17
PDB header: rna binding protein, transport proteinChain: A: PDB Molecule: signal recognition 54 kda protein;PDBTitle: structures of srp54 and srp19, the two proteins assembling2 the ribonucleic core of the signal recognition particle3 from the archaeon pyrococcus furiosus.
57 d1pkxa1
not modelled
19.9
17
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Inosicase58 c4a1oB_
not modelled
18.6
12
PDB header: transferase-hydrolaseChain: B: PDB Molecule: bifunctional purine biosynthesis protein purh;PDBTitle: crystal structure of mycobacterium tuberculosis purh complexed with2 aicar and a novel nucleotide cfair, at 2.48 a resolution.
59 d2a9pa1
not modelled
17.6
14
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related60 c3m6mF_
not modelled
17.5
26
PDB header: lyase/transferaseChain: F: PDB Molecule: sensory/regulatory protein rpfc;PDBTitle: crystal structure of rpff complexed with rec domain of rpfc
61 d1s8na_
not modelled
15.3
17
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related62 d1ys7a2
not modelled
14.9
11
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related63 c3cwoX_
not modelled
14.4
25
PDB header: de novo proteinChain: X: PDB Molecule: beta/alpha-barrel protein based on 1thf and 1tmy;PDBTitle: a beta/alpha-barrel built by the combination of fragments2 from different folds
64 c3oaaO_
not modelled
14.0
21
PDB header: hydrolase/transport proteinChain: O: PDB Molecule: atp synthase gamma chain;PDBTitle: structure of the e.coli f1-atp synthase inhibited by subunit epsilon
65 d1u0sy_
not modelled
14.0
22
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related66 c2nwqA_
not modelled
14.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: probable short-chain dehydrogenase;PDBTitle: short chain dehydrogenase from pseudomonas aeruginosa
67 d1g8fa3
not modelled
13.6
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ATP sulfurylase C-terminal domain68 c2vy9A_
not modelled
13.2
16
PDB header: gene regulationChain: A: PDB Molecule: anti-sigma-factor antagonist;PDBTitle: molecular architecture of the stressosome, a signal2 integration and transduction hub
69 c3qg5D_
not modelled
13.2
15
PDB header: hydrolaseChain: D: PDB Molecule: mre11;PDBTitle: the mre11:rad50 complex forms an atp dependent molecular clamp in dna2 double-strand break repair
70 c3dzdA_
not modelled
13.2
14
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigma54 activator ntrc4 in the inactive2 state
71 d2enda_
not modelled
12.6
63
Fold: T4 endonuclease VSuperfamily: T4 endonuclease VFamily: T4 endonuclease V72 d1q74a_
not modelled
12.6
16
Fold: LmbE-likeSuperfamily: LmbE-likeFamily: LmbE-like73 d2f99a1
not modelled
12.6
16
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: SnoaL-like polyketide cyclase74 c2qy9A_
not modelled
11.8
13
PDB header: protein transportChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of the ng+1 construct of the e. coli srp receptor2 ftsy
75 d2b7oa1
not modelled
11.8
19
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class-II DAHP synthetase76 d1c8ba_
not modelled
11.7
24
Fold: Phosphorylase/hydrolase-likeSuperfamily: HybD-likeFamily: Germination protease77 c3hv0A_
not modelled
11.6
10
PDB header: ligaseChain: A: PDB Molecule: tryptophanyl-trna synthetase;PDBTitle: tryptophanyl-trna synthetase from cryptosporidium parvum
78 c2q8uA_
not modelled
11.3
13
PDB header: hydrolaseChain: A: PDB Molecule: exonuclease, putative;PDBTitle: crystal structure of mre11 from thermotoga maritima msb8 (tm1635) at2 2.20 a resolution
79 c3lcmB_
not modelled
10.6
13
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of smu.1420 from streptococcus mutans ua159
80 c1iq0A_
not modelled
10.3
35
PDB header: ligaseChain: A: PDB Molecule: arginyl-trna synthetase;PDBTitle: thermus thermophilus arginyl-trna synthetase
81 c2j48A_
not modelled
10.1
11
PDB header: transferaseChain: A: PDB Molecule: two-component sensor kinase;PDBTitle: nmr structure of the pseudo-receiver domain of the cika2 protein.
82 c3t8yA_
not modelled
10.0
19
PDB header: hydrolaseChain: A: PDB Molecule: chemotaxis response regulator protein-glutamatePDBTitle: crystal structure of the response regulator domain of thermotoga2 maritima cheb
83 c2gwrA_
not modelled
9.9
12
PDB header: signaling proteinChain: A: PDB Molecule: dna-binding response regulator mtra;PDBTitle: crystal structure of the response regulator protein mtra from2 mycobacterium tuberculosis
84 c2g2qB_
not modelled
9.7
27
PDB header: oxidoreductaseChain: B: PDB Molecule: glutaredoxin-2;PDBTitle: the crystal structure of g4, the poxviral disulfide oxidoreductase2 essential for cytoplasmic disulfide bond formation
85 c3grcD_
not modelled
9.5
10
PDB header: transferaseChain: D: PDB Molecule: sensor protein, kinase;PDBTitle: crystal structure of a sensor protein from polaromonas sp.2 js666
86 c2xmoB_
not modelled
9.3
23
PDB header: hydrolaseChain: B: PDB Molecule: lmo2642 protein;PDBTitle: the crystal structure of lmo2642
87 c2og2A_
not modelled
9.3
16
PDB header: protein transportChain: A: PDB Molecule: putative signal recognition particle receptor;PDBTitle: crystal structure of chloroplast ftsy from arabidopsis2 thaliana
88 c2zwmA_
not modelled
9.1
15
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein yycf;PDBTitle: crystal structure of yycf receiver domain from bacillus2 subtilis
89 d1fs0g_
not modelled
8.9
15
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: ATP synthase (F1-ATPase), gamma subunitFamily: ATP synthase (F1-ATPase), gamma subunit90 c3a0rB_
not modelled
8.9
26
PDB header: transferaseChain: B: PDB Molecule: response regulator;PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
91 d1lpbb2
not modelled
8.8
44
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Pancreatic lipase, N-terminal domain92 d2qwxa1
not modelled
8.8
11
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase93 d1mb3a_
not modelled
8.7
15
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related94 c3cu5B_
not modelled
8.7
7
PDB header: transcription regulatorChain: B: PDB Molecule: two component transcriptional regulator, arac family;PDBTitle: crystal structure of a two component transcriptional regulator arac2 from clostridium phytofermentans isdg
95 c3bh1A_
not modelled
8.6
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0371 protein dip2346;PDBTitle: crystal structure of protein dip2346 from corynebacterium diphtheriae
96 c2qv0A_
not modelled
8.6
15
PDB header: transcriptionChain: A: PDB Molecule: protein mrke;PDBTitle: crystal structure of the response regulatory domain of2 protein mrke from klebsiella pneumoniae
97 d1dz3a_
not modelled
8.4
7
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related98 d1hska2
not modelled
8.4
23
Fold: Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domainSuperfamily: Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domainFamily: Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain99 c2xokG_
not modelled
8.4
33
PDB header: hydrolaseChain: G: PDB Molecule: atp synthase subunit gamma, mitochondrial;PDBTitle: refined structure of yeast f1c10 atpase complex to 3 a2 resolution