1 c3ed4A_
100.0
27
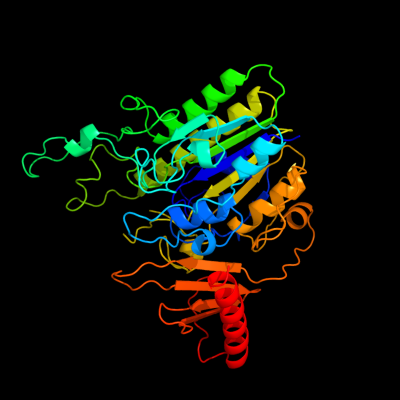
PDB header: transferaseChain: A: PDB Molecule: arylsulfatase;PDBTitle: crystal structure of putative arylsulfatase from escherichia coli
2 d1hdha_
100.0
29
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase3 c2qzuA_
100.0
23
PDB header: hydrolaseChain: A: PDB Molecule: putative sulfatase yidj;PDBTitle: crystal structure of the putative sulfatase yidj from bacteroides2 fragilis. northeast structural genomics consortium target bfr123
4 d1fsua_
100.0
27
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase5 d1p49a_
100.0
30
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase6 d1auka_
100.0
26
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase7 c2vqrA_
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: putative sulfatase;PDBTitle: crystal structure of a phosphonate monoester hydrolase2 from rhizobium leguminosarum: a new member of the3 alkaline phosphatase superfamily
8 c3b5qB_
100.0
24
PDB header: hydrolaseChain: B: PDB Molecule: putative sulfatase yidj;PDBTitle: crystal structure of a putative sulfatase (np_810509.1)2 from bacteroides thetaiotaomicron vpi-5482 at 2.40 a3 resolution
9 c3lxqB_
100.0
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein vp1736;PDBTitle: the crystal structure of a protein in the alkaline2 phosphatase superfamily from vibrio parahaemolyticus to3 1.95a
10 c2w8dB_
100.0
19
PDB header: transferaseChain: B: PDB Molecule: processed glycerol phosphate lipoteichoic acid synthase 2;PDBTitle: distinct and essential morphogenic functions for wall- and2 lipo-teichoic acids in bacillus subtilis
11 c2w5tA_
100.0
19
PDB header: transferaseChain: A: PDB Molecule: processed glycerol phosphate lipoteichoic acidPDBTitle: structure-based mechanism of lipoteichoic acid synthesis by2 staphylococcus aureus ltas.
12 c2zktB_
100.0
19
PDB header: isomeraseChain: B: PDB Molecule: 2,3-bisphosphoglycerate-independent phosphoglyceratePDBTitle: structure of ph0037 protein from pyrococcus horikoshii
13 c3m8yC_
100.0
16
PDB header: isomeraseChain: C: PDB Molecule: phosphopentomutase;PDBTitle: phosphopentomutase from bacillus cereus after glucose-1,6-bisphosphate2 activation
14 c3q3qA_
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase;PDBTitle: crystal structure of spap: an novel alkaline phosphatase from2 bacterium sphingomonas sp. strain bsar-1
15 c2gsoB_
100.0
24
PDB header: hydrolaseChain: B: PDB Molecule: phosphodiesterase-nucleotide pyrophosphatase;PDBTitle: structure of xac nucleotide2 pyrophosphatase/phosphodiesterase in complex with vanadate
16 c2i09A_
100.0
15
PDB header: isomeraseChain: A: PDB Molecule: phosphopentomutase;PDBTitle: crystal structure of putative phosphopentomutase from streptococcus2 mutans
17 d1o98a2
100.0
18
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, catalytic domain18 c3szzA_
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: phosphonoacetate hydrolase;PDBTitle: crystal structure of phosphonoacetate hydrolase from sinorhizobium2 meliloti 1021 in complex with acetate
19 d2i09a1
100.0
20
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: DeoB catalytic domain-like20 d1ei6a_
100.0
16
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Phosphonoacetate hydrolase21 c2xrgA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: crystal structure of autotaxin (enpp2) in complex with the2 ha155 boronic acid inhibitor
22 c2xr9A_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: crystal structure of autotaxin (enpp2)
23 c2d1gB_
not modelled
99.9
11
PDB header: hydrolaseChain: B: PDB Molecule: acid phosphatase;PDBTitle: structure of francisella tularensis acid phosphatase a (acpa) bound to2 orthovanadate
24 c1o98A_
not modelled
99.9
22
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independentPDBTitle: 1.4a crystal structure of phosphoglycerate mutase from2 bacillus stearothermophilus complexed with3 2-phosphoglycerate
25 c3igzB_
not modelled
99.8
14
PDB header: isomeraseChain: B: PDB Molecule: cofactor-independent phosphoglycerate mutase;PDBTitle: crystal structures of leishmania mexicana phosphoglycerate2 mutase at low cobalt concentration
26 d1y6va1
not modelled
99.6
19
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase27 c2iucB_
not modelled
99.6
14
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: structure of alkaline phosphatase from the antarctic2 bacterium tab5
28 c2w0yB_
not modelled
99.5
14
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: h.salinarum alkaline phosphatase
29 c2x98A_
not modelled
99.5
16
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase;PDBTitle: h.salinarum alkaline phosphatase
30 c1ew2A_
not modelled
99.5
17
PDB header: hydrolaseChain: A: PDB Molecule: phosphatase;PDBTitle: crystal structure of a human phosphatase
31 d1k7ha_
not modelled
99.5
17
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase32 d1zeda1
not modelled
99.5
17
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase33 c3a52A_
not modelled
99.4
14
PDB header: hydrolaseChain: A: PDB Molecule: cold-active alkaline phosphatase;PDBTitle: crystal structure of cold-active alkailne phosphatase from2 psychrophile shewanella sp.
34 c3e2dB_
not modelled
99.3
16
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: the 1.4 a crystal structure of the large and cold-active2 vibrio sp. alkaline phosphatase
35 c3iddA_
not modelled
95.8
30
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independentPDBTitle: cofactor-independent phosphoglycerate mutase from2 thermoplasma acidophilum dsm 1728
36 d1b4ub_
not modelled
59.3
24
Fold: Phosphorylase/hydrolase-likeSuperfamily: LigB-likeFamily: LigB-like37 d3cu0a1
not modelled
40.3
35
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: 1,3-glucuronyltransferase38 d2iw0a1
not modelled
33.7
34
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase39 c2d0jD_
not modelled
33.4
24
PDB header: transferaseChain: D: PDB Molecule: galactosylgalactosylxylosylprotein 3-beta-PDBTitle: crystal structure of human glcat-s apo form
40 c2r1fB_
not modelled
32.7
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: predicted aminodeoxychorismate lyase;PDBTitle: crystal structure of predicted aminodeoxychorismate lyase from2 escherichia coli
41 d1v82a_
not modelled
30.8
34
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: 1,3-glucuronyltransferase42 c3oaaO_
not modelled
25.4
6
PDB header: hydrolase/transport proteinChain: O: PDB Molecule: atp synthase gamma chain;PDBTitle: structure of the e.coli f1-atp synthase inhibited by subunit epsilon
43 d1xo1a2
not modelled
24.2
15
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: 5' to 3' exonuclease catalytic domain44 c2iw0A_
not modelled
24.2
36
PDB header: hydrolaseChain: A: PDB Molecule: chitin deacetylase;PDBTitle: structure of the chitin deacetylase from the fungal2 pathogen colletotrichum lindemuthianum
45 d1h7na_
not modelled
21.1
20
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)46 d1j33a_
not modelled
18.6
11
Fold: Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)Superfamily: Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)Family: Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)47 d1mm0a_
not modelled
18.0
6
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Insect defensins48 d1okga1
not modelled
17.9
15
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)49 d1dc1a_
not modelled
17.9
16
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease BsobI50 d1l5oa_
not modelled
16.9
20
Fold: Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)Superfamily: Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)Family: Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)51 d1skyb3
not modelled
16.5
33
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)52 d1tfra2
not modelled
15.2
19
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: 5' to 3' exonuclease catalytic domain53 d1fx0a3
not modelled
14.4
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)54 c3oaaC_
not modelled
14.1
19
PDB header: hydrolase/transport proteinChain: C: PDB Molecule: atp synthase subunit alpha;PDBTitle: structure of the e.coli f1-atp synthase inhibited by subunit epsilon
55 d1uzdc1
not modelled
13.9
19
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit56 c2vyoA_
not modelled
11.7
19
PDB header: hydrolaseChain: A: PDB Molecule: chitooligosaccharide deacetylase;PDBTitle: chitin deacetylase family member from encephalitozoon2 cuniculi
57 d1v7ba2
not modelled
11.7
21
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain58 c3k6sB_
not modelled
11.6
18
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-2;PDBTitle: structure of integrin alphaxbeta2 ectodomain
59 d2cz4a1
not modelled
11.6
19
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein60 d1l6sa_
not modelled
11.5
33
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)61 d1skye3
not modelled
11.1
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)62 c3k1tA_
not modelled
10.9
17
PDB header: ligaseChain: A: PDB Molecule: glutamate--cysteine ligase gsha;PDBTitle: crystal structure of putative gamma-glutamylcysteine synthetase2 (yp_546622.1) from methylobacillus flagellatus kt at 1.90 a3 resolution
63 d1ej7s_
not modelled
10.4
21
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit64 c1kmhA_
not modelled
10.3
15
PDB header: hydrolaseChain: A: PDB Molecule: atpase alpha subunit;PDBTitle: crystal structure of spinach chloroplast f1-atpase2 complexed with tentoxin
65 c3nrxB_
not modelled
10.2
11
PDB header: protein bindingChain: B: PDB Molecule: protein regulator of cytokinesis 1;PDBTitle: insights into anti-parallel microtubule crosslinking by prc1, a2 conserved non-motor microtubule binding protein
66 c2w6jG_
not modelled
10.1
25
PDB header: hydrolaseChain: G: PDB Molecule: atp synthase subunit gamma, mitochondrial;PDBTitle: low resolution structures of bovine mitochondrial f1-atpase2 during controlled dehydration: hydration state 5.
67 d2b8ea1
not modelled
9.8
32
Fold: HAD-likeSuperfamily: HAD-likeFamily: Meta-cation ATPase, catalytic domain P68 c2wl8D_
not modelled
9.0
24
PDB header: protein transportChain: D: PDB Molecule: peroxisomal biogenesis factor 19;PDBTitle: x-ray crystal structure of pex19p
69 d1gzga_
not modelled
8.9
26
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)70 c1w0jB_
not modelled
8.7
15
PDB header: hydrolaseChain: B: PDB Molecule: atp synthase alpha chain heart isoform,PDBTitle: beryllium fluoride inhibited bovine f1-atpase
71 d2c1ha1
not modelled
8.7
26
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)72 c2ihnA_
not modelled
8.7
16
PDB header: hydrolase/dnaChain: A: PDB Molecule: ribonuclease h;PDBTitle: co-crystal of bacteriophage t4 rnase h with a fork dna2 substrate
73 c3tixB_
not modelled
8.6
50
PDB header: gene regulation/protein bindingChain: B: PDB Molecule: chromo domain-containing protein 1;PDBTitle: crystal structure of the chp1-tas3 complex core
74 c1okgA_
not modelled
8.4
14
PDB header: transferaseChain: A: PDB Molecule: possible 3-mercaptopyruvate sulfurtransferase;PDBTitle: 3-mercaptopyruvate sulfurtransferase from leishmania major
75 c2qe7G_
not modelled
8.1
44
PDB header: hydrolaseChain: G: PDB Molecule: atp synthase subunit gamma;PDBTitle: crystal structure of the f1-atpase from the thermoalkaliphilic2 bacterium bacillus sp. ta2.a1
76 c2c61A_
not modelled
8.0
7
PDB header: hydrolaseChain: A: PDB Molecule: a-type atp synthase non-catalytic subunit b;PDBTitle: crystal structure of the non-catalytic b subunit of a-type2 atpase from m. mazei go1
77 c2pw0A_
not modelled
7.8
15
PDB header: unknown functionChain: A: PDB Molecule: prpf methylaconitate isomerase;PDBTitle: crystal structure of trans-aconitate bound to methylaconitate2 isomerase prpf from shewanella oneidensis
78 d2jdig1
not modelled
7.7
38
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: ATP synthase (F1-ATPase), gamma subunitFamily: ATP synthase (F1-ATPase), gamma subunit79 c3olhA_
not modelled
7.6
12
PDB header: transferaseChain: A: PDB Molecule: 3-mercaptopyruvate sulfurtransferase;PDBTitle: human 3-mercaptopyruvate sulfurtransferase
80 d8ruci_
not modelled
7.6
19
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit81 c3obkH_
not modelled
7.3
41
PDB header: lyaseChain: H: PDB Molecule: delta-aminolevulinic acid dehydratase;PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
82 c2qe7C_
not modelled
7.2
26
PDB header: hydrolaseChain: C: PDB Molecule: atp synthase subunit alpha;PDBTitle: crystal structure of the f1-atpase from the thermoalkaliphilic2 bacterium bacillus sp. ta2.a1
83 c2r9vA_
not modelled
7.1
22
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase subunit alpha;PDBTitle: crystal structure of atp synthase subunit alpha (tm1612) from2 thermotoga maritima at 2.10 a resolution
84 c2xokG_
not modelled
7.0
26
PDB header: hydrolaseChain: G: PDB Molecule: atp synthase subunit gamma, mitochondrial;PDBTitle: refined structure of yeast f1c10 atpase complex to 3 a2 resolution
85 c2fa5B_
not modelled
6.9
25
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator marr/emrr family;PDBTitle: the crystal structure of an unliganded multiple antibiotic-2 resistance repressor (marr) from xanthomonas campestris
86 c2w6fA_
not modelled
6.6
15
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase subunit alpha heart isoform,PDBTitle: low resolution structures of bovine mitochondrial f1-atpase2 during controlled dehydration: hydration state 2.
87 c2go4A_
not modelled
6.5
24
PDB header: hydrolaseChain: A: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] n-acetylglucosaminePDBTitle: crystal structure of aquifex aeolicus lpxc complexed with tu-514
88 c1u8cB_
not modelled
6.5
23
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: a novel adaptation of the integrin psi domain revealed from its2 crystal structure
89 c3p3vB_
not modelled
6.5
38
PDB header: transferaseChain: B: PDB Molecule: pts system, n-acetylgalactosamine-specific iib component;PDBTitle: crystal structure of a pts dependent n-acetyl-galactosamine-iib2 component (agav, spy_0631) from streptococcus pyogenes at 1.65 a3 resolution
90 d1szpb1
not modelled
6.5
14
Fold: SAM domain-likeSuperfamily: Rad51 N-terminal domain-likeFamily: DNA repair protein Rad51, N-terminal domain91 c3ov5A_
not modelled
5.9
13
PDB header: protein transportChain: A: PDB Molecule: uncharacterized protein;PDBTitle: atomic structure of the xanthomonas citri virb7 globular domain.
92 c3iydC_
not modelled
5.9
16
PDB header: transcription/dnaChain: C: PDB Molecule: dna-directed rna polymerase subunit beta;PDBTitle: three-dimensional em structure of an intact activator-dependent2 transcription initiation complex
93 c2eceA_
not modelled
5.8
4
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 462aa long hypothetical selenium-binding protein;PDBTitle: x-ray structure of hypothetical selenium-binding protein2 from sulfolobus tokodaii, st0059
94 d2jdia3
not modelled
5.5
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)95 c2vocA_
not modelled
5.5
29
PDB header: electron transportChain: A: PDB Molecule: thioredoxin;PDBTitle: thioredoxin a active site mutants form mixed disulfide2 dimers that resemble enzyme-substrate reaction3 intermediate
96 c3ippA_
not modelled
5.4
16
PDB header: transferaseChain: A: PDB Molecule: putative thiosulfate sulfurtransferase ynje;PDBTitle: crystal structure of sulfur-free ynje
97 d1qopb_
not modelled
5.4
11
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes98 d1g8fa3
not modelled
5.3
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ATP sulfurylase C-terminal domain99 d1tg7a2
not modelled
5.2
25
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Beta-galactosidase LacA, domains 4 and 5