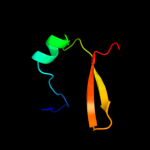
| 1 | c2w1tB_ |
|
 |
98.6 |
21 |
PDB header:transcription
Chain: B: PDB Molecule:stage v sporulation protein t;
PDBTitle: crystal structure of b. subtilis spovt
|
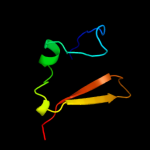
| 2 | c2l66B_ |
|
 |
98.2 |
29 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, abrb family;
PDBTitle: the dna-recognition fold of sso7c4 suggests a new member of spovt-abrb2 superfamily from archaea.
|
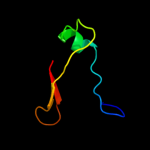
| 3 | c2ro5B_ |
|
 |
97.5 |
23 |
PDB header:transcription
Chain: B: PDB Molecule:stage v sporulation protein t;
PDBTitle: rdc-refined solution structure of the n-terminal dna2 recognition domain of the bacillus subtilis transition-3 state regulator spovt
|
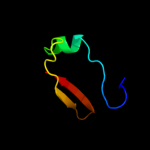
| 4 | d2fy9a1 |
|
 |
97.2 |
26 |
Fold:Double-split beta-barrel
Superfamily:AbrB/MazE/MraZ-like
Family:AbrB N-terminal domain-like |
| 5 | d1yfba1 |
|
 |
97.1 |
21 |
Fold:Double-split beta-barrel
Superfamily:AbrB/MazE/MraZ-like
Family:AbrB N-terminal domain-like |
| 6 | c2glwA_ |
|
 |
97.0 |
27 |
PDB header:transcription
Chain: A: PDB Molecule:92aa long hypothetical protein;
PDBTitle: the solution structure of phs018 from pyrococcus horikoshii
|
| 7 | d1ub4c_ |
|
 |
95.6 |
22 |
Fold:Double-split beta-barrel
Superfamily:AbrB/MazE/MraZ-like
Family:Kis/PemI addiction antidote |
| 8 | d1mvfd_ |
|
 |
94.9 |
24 |
Fold:Double-split beta-barrel
Superfamily:AbrB/MazE/MraZ-like
Family:Kis/PemI addiction antidote |
| 9 | d1n0ea_ |
|
 |
90.6 |
14 |
Fold:Double-split beta-barrel
Superfamily:AbrB/MazE/MraZ-like
Family:Hypothetical protein MraZ |
| 10 | c1n0fF_ |
|
 |
90.4 |
14 |
PDB header:biosynthetic protein
Chain: F: PDB Molecule:protein mraz;
PDBTitle: crystal structure of a cell division and cell wall2 biosynthesis protein upf0040 from mycoplasma pneumoniae:3 indication of a novel fold with a possible new conserved4 sequence motif
|
| 11 | d2d9ra1 |
|
 |
84.6 |
32 |
Fold:Double-split beta-barrel
Superfamily:AF2212/PG0164-like
Family:PG0164-like |
| 12 | d1ylea1 |
|
 |
83.7 |
14 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:AstA-like |
| 13 | d2vbua1 |
|
 |
78.2 |
41 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin kinase-like
Family:CTP-dependent riboflavin kinase-like |
| 14 | c2c45F_ |
|
 |
76.0 |
20 |
PDB header:lyase
Chain: F: PDB Molecule:aspartate 1-decarboxylase precursor;
PDBTitle: native precursor of pyruvoyl dependent aspartate2 decarboxylase
|
| 15 | c3o27B_ |
|
 |
73.8 |
29 |
PDB header:dna binding protein
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: the crystal structure of c68 from the hybrid virus-plasmid pssvx
|
| 16 | d1ppya_ |
|
 |
71.6 |
18 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Pyruvoyl dependent aspartate decarboxylase, ADC |
| 17 | c1pt1B_ |
|
 |
71.6 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:aspartate 1-decarboxylase;
PDBTitle: unprocessed pyruvoyl dependent aspartate decarboxylase with histidine2 11 mutated to alanine
|
| 18 | c2l55A_ |
|
 |
67.6 |
27 |
PDB header:metal binding protein
Chain: A: PDB Molecule:silb,silver efflux protein, mfp component of the three
PDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
|
| 19 | c1zeqX_ |
|
 |
67.1 |
19 |
PDB header:metal binding protein
Chain: X: PDB Molecule:cation efflux system protein cusf;
PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
|
| 20 | c2pjhB_ |
|
 |
61.1 |
16 |
PDB header:transport protein
Chain: B: PDB Molecule:transitional endoplasmic reticulum atpase;
PDBTitle: strctural model of the p97 n domain- npl4 ubd complex
|
| 21 | d2p19a1 |
|
not modelled |
55.5 |
17 |
Fold:Chorismate lyase-like
Superfamily:Chorismate lyase-like
Family:UTRA domain |
| 22 | c2kdoA_ |
|
not modelled |
49.4 |
18 |
PDB header:rna binding protein
Chain: A: PDB Molecule:ribosome maturation protein sbds;
PDBTitle: structure of the human shwachman-bodian-diamond syndrome protein, sbds
|
| 23 | d2c4ka1 |
|
not modelled |
48.4 |
11 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 24 | d2fa1a1 |
|
not modelled |
48.3 |
14 |
Fold:Chorismate lyase-like
Superfamily:Chorismate lyase-like
Family:UTRA domain |
| 25 | c3ll7A_ |
|
not modelled |
47.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:putative methyltransferase;
PDBTitle: crystal structure of putative methyltransferase pg_1098 from2 porphyromonas gingivalis w83
|
| 26 | d1u9ya1 |
|
not modelled |
45.9 |
16 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 27 | d2ikka1 |
|
not modelled |
44.2 |
17 |
Fold:Chorismate lyase-like
Superfamily:Chorismate lyase-like
Family:UTRA domain |
| 28 | c2ra5A_ |
|
not modelled |
44.0 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of the putative transcriptional regulator2 from streptomyces coelicolor
|
| 29 | d2ra5a1 |
|
not modelled |
44.0 |
19 |
Fold:Chorismate lyase-like
Superfamily:Chorismate lyase-like
Family:UTRA domain |
| 30 | d1cz5a1 |
|
not modelled |
42.1 |
20 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Cdc48 N-terminal domain-like |
| 31 | d3cnva1 |
|
not modelled |
41.8 |
24 |
Fold:Chorismate lyase-like
Superfamily:Chorismate lyase-like
Family:UTRA domain |
| 32 | c1zc1A_ |
|
not modelled |
41.5 |
24 |
PDB header:protein turnover
Chain: A: PDB Molecule:ubiquitin fusion degradation protein 1;
PDBTitle: ufd1 exhibits the aaa-atpase fold with two distinct2 ubiquitin interaction sites
|
| 33 | d2pkha1 |
|
not modelled |
40.8 |
12 |
Fold:Chorismate lyase-like
Superfamily:Chorismate lyase-like
Family:UTRA domain |
| 34 | c3pjyB_ |
|
not modelled |
40.5 |
25 |
PDB header:transcription regulator
Chain: B: PDB Molecule:hypothetical signal peptide protein;
PDBTitle: crystal structure of a putative transcription regulator (r01717) from2 sinorhizobium meliloti 1021 at 1.55 a resolution
|
| 35 | c3m7aA_ |
|
not modelled |
39.7 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of saro_0823 (yp_496102.1) a protein of2 unknown function from novosphingobium aromaticivorans dsm3 12444 at 1.22 a resolution
|
| 36 | d3bwga2 |
|
not modelled |
36.8 |
20 |
Fold:Chorismate lyase-like
Superfamily:Chorismate lyase-like
Family:UTRA domain |
| 37 | d1dkua1 |
|
not modelled |
35.4 |
8 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 38 | c1cz5A_ |
|
not modelled |
34.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:vcp-like atpase;
PDBTitle: nmr structure of vat-n: the n-terminal domain of vat (vcp-2 like atpase of thermoplasma)
|
| 39 | d1vlfm1 |
|
not modelled |
33.4 |
28 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 40 | c1zzvA_ |
|
not modelled |
33.4 |
15 |
PDB header:membrane protein, metal transport
Chain: A: PDB Molecule:iron(iii) dicitrate transport protein feca;
PDBTitle: solution nmr structure of the periplasmic signaling domain2 of the outer membrane iron transporter feca from3 escherichia coli.
|
| 41 | d2axti1 |
|
not modelled |
32.5 |
36 |
Fold:Single transmembrane helix
Superfamily:Photosystem II reaction center protein I, PsbI
Family:PsbI-like |
| 42 | c2yujA_ |
|
not modelled |
31.3 |
16 |
PDB header:protein binding
Chain: A: PDB Molecule:ubiquitin fusion degradation 1-like;
PDBTitle: solution structure of human ubiquitin fusion degradation2 protein 1 homolog ufd1
|
| 43 | c2ki8A_ |
|
not modelled |
30.5 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tungsten formylmethanofuran dehydrogenase,
PDBTitle: solution nmr structure of tungsten formylmethanofuran2 dehydrogenase subunit d from archaeoglobus fulgidus,3 northeast structural genomics consortium target att7
|
| 44 | c2k4vA_ |
|
not modelled |
30.5 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein pa1076;
PDBTitle: solution structure of uncharacterized protein pa1076 from2 pseudomonas aeruginosa. northeast structural genomics3 consortium (nesg) target pat3, ontario center for4 structural proteomics target pa1076 .
|
| 45 | d1zq1a1 |
|
not modelled |
28.3 |
14 |
Fold:Sm-like fold
Superfamily:GatD N-terminal domain-like
Family:GatD N-terminal domain-like |
| 46 | d1we0a1 |
|
not modelled |
27.6 |
12 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 47 | d2fug21 |
|
not modelled |
24.8 |
24 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:NQO2-like |
| 48 | d2f23a2 |
|
not modelled |
24.6 |
21 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:GreA transcript cleavage factor, C-terminal domain |
| 49 | c2jwlB_ |
|
not modelled |
24.6 |
14 |
PDB header:membrane protein
Chain: B: PDB Molecule:protein tolr;
PDBTitle: solution structure of periplasmic domain of tolr from h.2 influenzae with saxs data
|
| 50 | c3lheA_ |
|
not modelled |
23.7 |
23 |
PDB header:transcription regulator
Chain: A: PDB Molecule:gntr family transcriptional regulator;
PDBTitle: the crystal structure of the c-terminal domain of a gntr2 family transcriptional regulator from bacillus anthracis3 str. sterne
|
| 51 | d1yexa1 |
|
not modelled |
23.4 |
16 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 52 | c2oqkA_ |
|
not modelled |
23.4 |
14 |
PDB header:translation
Chain: A: PDB Molecule:putative translation initiation factor eif-1a;
PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
|
| 53 | c1q6xA_ |
|
not modelled |
23.0 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:choline o-acetyltransferase;
PDBTitle: crystal structure of rat choline acetyltransferase
|
| 54 | d1u0la1 |
|
not modelled |
23.0 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 55 | d1zvvj1 |
|
not modelled |
22.1 |
15 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 56 | c2c4kD_ |
|
not modelled |
22.0 |
11 |
PDB header:regulatory protein
Chain: D: PDB Molecule:phosphoribosyl pyrophosphate synthetase-
PDBTitle: crystal structure of human phosphoribosylpyrophosphate2 synthetase-associated protein 39 (pap39)
|
| 57 | c3hvzB_ |
|
not modelled |
21.7 |
22 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the tgs domain of the clolep_03100 protein from2 clostridium leptum, northeast structural genomics consortium target3 qlr13a
|
| 58 | d1qr5a_ |
|
not modelled |
21.7 |
13 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 59 | d2ifea_ |
|
not modelled |
21.6 |
14 |
Fold:IF3-like
Superfamily:Translation initiation factor IF3, C-terminal domain
Family:Translation initiation factor IF3, C-terminal domain |
| 60 | d1ka5a_ |
|
not modelled |
21.5 |
13 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 61 | c1t95A_ |
|
not modelled |
21.5 |
12 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein af0491;
PDBTitle: crystal structure of the shwachman-bodian-diamond syndrome2 protein orthologue from archaeoglobus fulgidus
|
| 62 | d3d31a1 |
|
not modelled |
20.8 |
18 |
Fold:OB-fold
Superfamily:MOP-like
Family:ABC-transporter additional domain |
| 63 | c3hfiA_ |
|
not modelled |
20.4 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative regulator;
PDBTitle: the crystal structure of the putative regulator from escherichia coli2 cft073
|
| 64 | c3dahB_ |
|
not modelled |
20.3 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: 2.3 a crystal structure of ribose-phosphate pyrophosphokinase from2 burkholderia pseudomallei
|
| 65 | c3ho6B_ |
|
not modelled |
19.9 |
16 |
PDB header:toxin
Chain: B: PDB Molecule:toxin a;
PDBTitle: structure-function analysis of inositol hexakisphosphate-2 induced autoprocessing in clostridium difficile toxin a
|
| 66 | c3f8lC_ |
|
not modelled |
19.7 |
11 |
PDB header:transcription
Chain: C: PDB Molecule:hth-type transcriptional repressor phnf;
PDBTitle: crystal structure of the effector domain of phnf from mycobacterium2 smegmatis
|
| 67 | c2wbmA_ |
|
not modelled |
19.4 |
16 |
PDB header:rna-binding protein
Chain: A: PDB Molecule:ribosome maturation protein sdo1 homolog;
PDBTitle: crystal structure of mthsbds, the homologue of the2 shwachman-bodian-diamond syndrome protein in the3 euriarchaeon methanothermobacter thermautotrophicus
|
| 68 | c3ougA_ |
|
not modelled |
19.4 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:aspartate 1-decarboxylase;
PDBTitle: crystal structure of cleaved l-aspartate-alpha-decarboxylase from2 francisella tularensis
|
| 69 | d1mo1a_ |
|
not modelled |
19.4 |
17 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 70 | d1ptfa_ |
|
not modelled |
19.2 |
15 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 71 | d2hpra_ |
|
not modelled |
18.8 |
17 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 72 | c3bmbB_ |
|
not modelled |
18.7 |
36 |
PDB header:rna binding protein
Chain: B: PDB Molecule:regulator of nucleoside diphosphate kinase;
PDBTitle: crystal structure of a new rna polymerase interacting2 protein
|
| 73 | d1hr0w_ |
|
not modelled |
18.6 |
35 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 74 | d1y7qa1 |
|
not modelled |
18.4 |
19 |
Fold:Acyl carrier protein-like
Superfamily:Retrovirus capsid dimerization domain-like
Family:SCAN domain |
| 75 | d2jioa1 |
|
not modelled |
17.6 |
16 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 76 | d1jt8a_ |
|
not modelled |
17.5 |
9 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 77 | d1ihra_ |
|
not modelled |
16.8 |
19 |
Fold:TolA/TonB C-terminal domain
Superfamily:TolA/TonB C-terminal domain
Family:TonB |
| 78 | d1g8ka1 |
|
not modelled |
16.8 |
13 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 79 | d2eifa2 |
|
not modelled |
16.5 |
37 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 80 | c3zvkG_ |
|
not modelled |
16.5 |
13 |
PDB header:antitoxin/toxin/dna
Chain: G: PDB Molecule:antitoxin of toxin-antitoxin system vapb;
PDBTitle: crystal structure of vapbc2 from rickettsia felis bound to2 a dna fragment from their promoter
|
| 81 | c3le1B_ |
|
not modelled |
16.4 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:phosphotransferase system, hpr-related proteins;
PDBTitle: crystal structure of apohpr monomer from thermoanaerobacter2 tengcongensis
|
| 82 | c3lotC_ |
|
not modelled |
16.3 |
39 |
PDB header:structure genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein of unknown function (np_070038.1) from2 archaeoglobus fulgidus at 1.89 a resolution
|
| 83 | c1u9yD_ |
|
not modelled |
16.3 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: crystal structure of phosphoribosyl diphosphate synthase2 from methanocaldococcus jannaschii
|
| 84 | c3i4oA_ |
|
not modelled |
16.1 |
26 |
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor if-1;
PDBTitle: crystal structure of translation initiation factor 1 from2 mycobacterium tuberculosis
|
| 85 | c3efhB_ |
|
not modelled |
15.9 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase 1;
PDBTitle: crystal structure of human phosphoribosyl pyrophosphate2 synthetase 1
|
| 86 | d1kqfa1 |
|
not modelled |
15.8 |
12 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 87 | c1h0hA_ |
|
not modelled |
15.5 |
13 |
PDB header:dehydrogenase
Chain: A: PDB Molecule:formate dehydrogenase (large subunit);
PDBTitle: tungsten containing formate dehydrogenase from2 desulfovibrio gigas
|
| 88 | d1kbla3 |
|
not modelled |
15.3 |
17 |
Fold:ATP-grasp
Superfamily:Glutathione synthetase ATP-binding domain-like
Family:Pyruvate phosphate dikinase, N-terminal domain |
| 89 | d2iv2x1 |
|
not modelled |
15.2 |
10 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 90 | c2pn0D_ |
|
not modelled |
14.6 |
38 |
PDB header:transcription
Chain: D: PDB Molecule:prokaryotic transcription elongation factor
PDBTitle: prokaryotic transcription elongation factor grea/greb from2 nitrosomonas europaea
|
| 91 | c1dkrB_ |
|
not modelled |
14.6 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosyl pyrophosphate synthetase;
PDBTitle: crystal structures of bacillus subtilis phosphoribosylpyrophosphate2 synthetase: molecular basis of allosteric inhibition and activation.
|
| 92 | d1h9ma2 |
|
not modelled |
14.5 |
32 |
Fold:OB-fold
Superfamily:MOP-like
Family:BiMOP, duplicated molybdate-binding domain |
| 93 | d1t9ha1 |
|
not modelled |
14.5 |
14 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 94 | c3lpnB_ |
|
not modelled |
14.5 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: crystal structure of the phosphoribosylpyrophosphate (prpp) synthetase2 from thermoplasma volcanium in complex with an atp analog (ampcpp).
|
| 95 | d2nzul1 |
|
not modelled |
14.5 |
18 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 96 | c3e02A_ |
|
not modelled |
14.4 |
28 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849;
PDBTitle: crystal structure of a duf849 family protein (bxe_c0271) from2 burkholderia xenovorans lb400 at 1.90 a resolution
|
| 97 | c1h9mB_ |
|
not modelled |
14.3 |
27 |
PDB header:binding protein
Chain: B: PDB Molecule:molybdenum-binding-protein;
PDBTitle: two crystal structures of the cytoplasmic molybdate-binding2 protein modg suggest a novel cooperative binding mechanism3 and provide insights into ligand-binding specificity.4 peg-grown form with molybdate bound
|
| 98 | c1vlfQ_ |
|
not modelled |
14.2 |
28 |
PDB header:oxidoreductase
Chain: Q: PDB Molecule:pyrogallol hydroxytransferase large subunit;
PDBTitle: crystal structure of pyrogallol-phloroglucinol2 transhydroxylase from pelobacter acidigallici complexed3 with inhibitor 1,2,4,5-tetrahydroxy-benzene
|
| 99 | d2etna2 |
|
not modelled |
13.9 |
21 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:GreA transcript cleavage factor, C-terminal domain |

