| 1 | c2w8dB_ |
|
 |
100.0 |
15 |
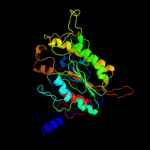
PDB header:transferase
Chain: B: PDB Molecule:processed glycerol phosphate lipoteichoic acid synthase 2;
PDBTitle: distinct and essential morphogenic functions for wall- and2 lipo-teichoic acids in bacillus subtilis
|
| 2 | c2w5tA_ |
|
 |
100.0 |
18 |
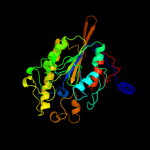
PDB header:transferase
Chain: A: PDB Molecule:processed glycerol phosphate lipoteichoic acid
PDBTitle: structure-based mechanism of lipoteichoic acid synthesis by2 staphylococcus aureus ltas.
|
| 3 | c3lxqB_ |
|
 |
100.0 |
17 |
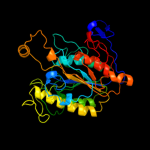
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein vp1736;
PDBTitle: the crystal structure of a protein in the alkaline2 phosphatase superfamily from vibrio parahaemolyticus to3 1.95a
|
| 4 | c2qzuA_ |
|
 |
100.0 |
18 |
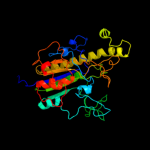
PDB header:hydrolase
Chain: A: PDB Molecule:putative sulfatase yidj;
PDBTitle: crystal structure of the putative sulfatase yidj from bacteroides2 fragilis. northeast structural genomics consortium target bfr123
|
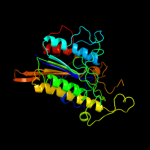
| 5 | c3ed4A_ |
|
 |
100.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:arylsulfatase;
PDBTitle: crystal structure of putative arylsulfatase from escherichia coli
|
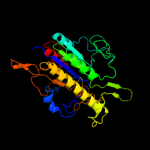
| 6 | d1fsua_ |
|
 |
100.0 |
10 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
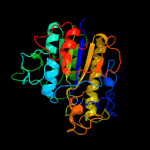
| 7 | d1auka_ |
|
 |
100.0 |
12 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
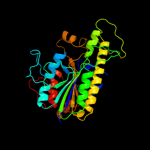
| 8 | c3b5qB_ |
|
 |
100.0 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative sulfatase yidj;
PDBTitle: crystal structure of a putative sulfatase (np_810509.1)2 from bacteroides thetaiotaomicron vpi-5482 at 2.40 a3 resolution
|
| 9 | d1hdha_ |
|
 |
100.0 |
16 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
| 10 | c2vqrA_ |
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative sulfatase;
PDBTitle: crystal structure of a phosphonate monoester hydrolase2 from rhizobium leguminosarum: a new member of the3 alkaline phosphatase superfamily
|
| 11 | d1p49a_ |
|
 |
100.0 |
16 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
| 12 | d1o98a2 |
|
 |
100.0 |
13 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, catalytic domain |
| 13 | d2i09a1 |
|
 |
99.9 |
14 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:DeoB catalytic domain-like |
| 14 | c2zktB_ |
|
 |
99.9 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:2,3-bisphosphoglycerate-independent phosphoglycerate
PDBTitle: structure of ph0037 protein from pyrococcus horikoshii
|
| 15 | c3m8yC_ |
|
 |
99.9 |
18 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphopentomutase;
PDBTitle: phosphopentomutase from bacillus cereus after glucose-1,6-bisphosphate2 activation
|
| 16 | c2gsoB_ |
|
 |
99.9 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:phosphodiesterase-nucleotide pyrophosphatase;
PDBTitle: structure of xac nucleotide2 pyrophosphatase/phosphodiesterase in complex with vanadate
|
| 17 | c3q3qA_ |
|
 |
99.9 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:alkaline phosphatase;
PDBTitle: crystal structure of spap: an novel alkaline phosphatase from2 bacterium sphingomonas sp. strain bsar-1
|
| 18 | c2i09A_ |
|
 |
99.8 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphopentomutase;
PDBTitle: crystal structure of putative phosphopentomutase from streptococcus2 mutans
|
| 19 | c3szzA_ |
|
 |
99.8 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonoacetate hydrolase;
PDBTitle: crystal structure of phosphonoacetate hydrolase from sinorhizobium2 meliloti 1021 in complex with acetate
|
| 20 | c2xr9A_ |
|
 |
99.8 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: crystal structure of autotaxin (enpp2)
|
| 21 | c2xrgA_ |
|
not modelled |
99.8 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: crystal structure of autotaxin (enpp2) in complex with the2 ha155 boronic acid inhibitor
|
| 22 | d1ei6a_ |
|
not modelled |
99.8 |
16 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Phosphonoacetate hydrolase |
| 23 | c1o98A_ |
|
not modelled |
99.7 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-bisphosphoglycerate-independent
PDBTitle: 1.4a crystal structure of phosphoglycerate mutase from2 bacillus stearothermophilus complexed with3 2-phosphoglycerate
|
| 24 | c3igzB_ |
|
not modelled |
99.6 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:cofactor-independent phosphoglycerate mutase;
PDBTitle: crystal structures of leishmania mexicana phosphoglycerate2 mutase at low cobalt concentration
|
| 25 | c2d1gB_ |
|
not modelled |
99.5 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:acid phosphatase;
PDBTitle: structure of francisella tularensis acid phosphatase a (acpa) bound to2 orthovanadate
|
| 26 | c2iucB_ |
|
not modelled |
99.2 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:alkaline phosphatase;
PDBTitle: structure of alkaline phosphatase from the antarctic2 bacterium tab5
|
| 27 | d1y6va1 |
|
not modelled |
99.1 |
14 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Alkaline phosphatase |
| 28 | c1ew2A_ |
|
not modelled |
99.0 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphatase;
PDBTitle: crystal structure of a human phosphatase
|
| 29 | d1zeda1 |
|
not modelled |
99.0 |
14 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Alkaline phosphatase |
| 30 | d1k7ha_ |
|
not modelled |
98.8 |
15 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Alkaline phosphatase |
| 31 | c3a52A_ |
|
not modelled |
98.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:cold-active alkaline phosphatase;
PDBTitle: crystal structure of cold-active alkailne phosphatase from2 psychrophile shewanella sp.
|
| 32 | c2w0yB_ |
|
not modelled |
98.7 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:alkaline phosphatase;
PDBTitle: h.salinarum alkaline phosphatase
|
| 33 | c2x98A_ |
|
not modelled |
98.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:alkaline phosphatase;
PDBTitle: h.salinarum alkaline phosphatase
|
| 34 | c3e2dB_ |
|
not modelled |
98.2 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:alkaline phosphatase;
PDBTitle: the 1.4 a crystal structure of the large and cold-active2 vibrio sp. alkaline phosphatase
|
| 35 | c3iddA_ |
|
not modelled |
95.4 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-bisphosphoglycerate-independent
PDBTitle: cofactor-independent phosphoglycerate mutase from2 thermoplasma acidophilum dsm 1728
|
| 36 | d1b4ub_ |
|
not modelled |
68.2 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:LigB-like
Family:LigB-like |
| 37 | d1s1qa_ |
|
not modelled |
67.1 |
21 |
Fold:UBC-like
Superfamily:UBC-like
Family:UEV domain |
| 38 | d1s95a_ |
|
not modelled |
41.2 |
18 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 39 | c2xmoB_ |
|
not modelled |
41.0 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:lmo2642 protein;
PDBTitle: the crystal structure of lmo2642
|
| 40 | d1uzdc1 |
|
not modelled |
33.2 |
17 |
Fold:RuBisCO, small subunit
Superfamily:RuBisCO, small subunit
Family:RuBisCO, small subunit |
| 41 | d1l5oa_ |
|
not modelled |
31.7 |
40 |
Fold:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Superfamily:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Family:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) |
| 42 | d1j33a_ |
|
not modelled |
31.2 |
27 |
Fold:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Superfamily:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Family:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) |
| 43 | d1ej7s_ |
|
not modelled |
27.9 |
17 |
Fold:RuBisCO, small subunit
Superfamily:RuBisCO, small subunit
Family:RuBisCO, small subunit |
| 44 | d8ruci_ |
|
not modelled |
23.4 |
17 |
Fold:RuBisCO, small subunit
Superfamily:RuBisCO, small subunit
Family:RuBisCO, small subunit |
| 45 | c3uoaB_ |
|
not modelled |
20.7 |
24 |
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:mucosa-associated lymphoid tissue lymphoma translocation
PDBTitle: crystal structure of the malt1 paracaspase (p21 form)
|
| 46 | c3bijC_ |
|
not modelled |
19.4 |
19 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein gsu0716;
PDBTitle: crystal structure of protein gsu0716 from geobacter2 sulfurreducens. northeast structural genomics target gsr13
|
| 47 | d2b8ea1 |
|
not modelled |
18.6 |
18 |
Fold:HAD-like
Superfamily:HAD-like
Family:Meta-cation ATPase, catalytic domain P |
| 48 | c2xokG_ |
|
not modelled |
17.0 |
15 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: refined structure of yeast f1c10 atpase complex to 3 a2 resolution
|
| 49 | d1xo1a2 |
|
not modelled |
16.9 |
12 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 50 | d1fs0g_ |
|
not modelled |
16.8 |
21 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:ATP synthase (F1-ATPase), gamma subunit
Family:ATP synthase (F1-ATPase), gamma subunit |
| 51 | d1tfra2 |
|
not modelled |
16.6 |
16 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 52 | d2nvpa1 |
|
not modelled |
16.3 |
23 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:CPF0428-like |
| 53 | d2hkja2 |
|
not modelled |
14.8 |
20 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:DNA gyrase/MutL, second domain |
| 54 | d1uf3a_ |
|
not modelled |
14.6 |
9 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TT1561-like |
| 55 | c1jaeA_ |
|
not modelled |
14.4 |
16 |
PDB header:glycosidase
Chain: A: PDB Molecule:alpha-amylase;
PDBTitle: structure of tenebrio molitor larval alpha-amylase
|
| 56 | c3oaaO_ |
|
not modelled |
14.3 |
17 |
PDB header:hydrolase/transport protein
Chain: O: PDB Molecule:atp synthase gamma chain;
PDBTitle: structure of the e.coli f1-atp synthase inhibited by subunit epsilon
|
| 57 | c2ihnA_ |
|
not modelled |
13.9 |
12 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:ribonuclease h;
PDBTitle: co-crystal of bacteriophage t4 rnase h with a fork dna2 substrate
|
| 58 | d1yj5a1 |
|
not modelled |
12.8 |
21 |
Fold:HAD-like
Superfamily:HAD-like
Family:phosphatase domain of polynucleotide kinase |
| 59 | d2bdua1 |
|
not modelled |
12.5 |
14 |
Fold:HAD-like
Superfamily:HAD-like
Family:Pyrimidine 5'-nucleotidase (UMPH-1) |
| 60 | d1usha2 |
|
not modelled |
11.7 |
26 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
| 61 | d1wdds_ |
|
not modelled |
11.6 |
19 |
Fold:RuBisCO, small subunit
Superfamily:RuBisCO, small subunit
Family:RuBisCO, small subunit |
| 62 | d1cmwa2 |
|
not modelled |
11.5 |
8 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 63 | c2w6jG_ |
|
not modelled |
11.0 |
22 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: low resolution structures of bovine mitochondrial f1-atpase2 during controlled dehydration: hydration state 5.
|
| 64 | d1qopb_ |
|
not modelled |
10.1 |
24 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 65 | d1a3xa3 |
|
not modelled |
10.1 |
19 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
| 66 | c3kccA_ |
|
not modelled |
9.9 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:catabolite gene activator;
PDBTitle: crystal structure of d138l mutant of catabolite gene activator protein
|
| 67 | d2hy1a1 |
|
not modelled |
9.9 |
20 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:GpdQ-like |
| 68 | c2hy1A_ |
|
not modelled |
9.9 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:rv0805;
PDBTitle: crystal structure of rv0805
|
| 69 | c2qe7G_ |
|
not modelled |
9.7 |
37 |
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma;
PDBTitle: crystal structure of the f1-atpase from the thermoalkaliphilic2 bacterium bacillus sp. ta2.a1
|
| 70 | c3ib7A_ |
|
not modelled |
9.2 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:icc protein;
PDBTitle: crystal structure of full length rv0805
|
| 71 | d2fiqa1 |
|
not modelled |
9.0 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:GatZ-like |
| 72 | d2f9zc1 |
|
not modelled |
8.9 |
23 |
Fold:CNF1/YfiH-like putative cysteine hydrolases
Superfamily:CNF1/YfiH-like putative cysteine hydrolases
Family:CheD-like |
| 73 | d1fyea_ |
|
not modelled |
8.7 |
10 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Aspartyl dipeptidase PepE |
| 74 | c3qfnA_ |
|
not modelled |
8.7 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of streptococcal asymmetric ap4a hydrolase and2 phosphodiesterase spr1479/saph in complex with inorganic phosphate
|
| 75 | c3e20C_ |
|
not modelled |
8.7 |
11 |
PDB header:translation
Chain: C: PDB Molecule:eukaryotic peptide chain release factor subunit 1;
PDBTitle: crystal structure of s.pombe erf1/erf3 complex
|
| 76 | c2pr7A_ |
|
not modelled |
8.6 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:haloacid dehalogenase/epoxide hydrolase family;
PDBTitle: crystal structure of uncharacterized protein (np_599989.1) from2 corynebacterium glutamicum atcc 13032 kitasato at 1.44 a resolution
|
| 77 | c1zreB_ |
|
not modelled |
8.3 |
15 |
PDB header:gene regulation/dna
Chain: B: PDB Molecule:catabolite gene activator;
PDBTitle: 4 crystal structures of cap-dna with all base-pair2 substitutions at position 6, cap-[6g;17c]icap38 dna
|
| 78 | d1jaea2 |
|
not modelled |
8.2 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 79 | c3qg5D_ |
|
not modelled |
8.2 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:mre11;
PDBTitle: the mre11:rad50 complex forms an atp dependent molecular clamp in dna2 double-strand break repair
|
| 80 | c3e37B_ |
|
not modelled |
8.1 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:protein farnesyltransferase subunit beta;
PDBTitle: protein farnesyltransferase complexed with bisubstrate2 ethylenediamine scaffold inhibitor 5
|
| 81 | d1wpga2 |
|
not modelled |
8.1 |
18 |
Fold:HAD-like
Superfamily:HAD-like
Family:Meta-cation ATPase, catalytic domain P |
| 82 | c2zqeA_ |
|
not modelled |
8.1 |
20 |
PDB header:dna binding protein
Chain: A: PDB Molecule:muts2 protein;
PDBTitle: crystal structure of the smr domain of thermus thermophilus muts2
|
| 83 | c2agaA_ |
|
not modelled |
8.0 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:machado-joseph disease protein 1;
PDBTitle: de-ubiquitinating function of ataxin-3: insights from the2 solution structure of the josephin domain
|
| 84 | c3t1iC_ |
|
not modelled |
8.0 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:double-strand break repair protein mre11a;
PDBTitle: crystal structure of human mre11: understanding tumorigenic mutations
|
| 85 | d2h6fb1 |
|
not modelled |
7.9 |
14 |
Fold:alpha/alpha toroid
Superfamily:Terpenoid cyclases/Protein prenyltransferases
Family:Protein prenyltransferases |
| 86 | d1okga1 |
|
not modelled |
7.8 |
8 |
Fold:Rhodanese/Cell cycle control phosphatase
Superfamily:Rhodanese/Cell cycle control phosphatase
Family:Multidomain sulfurtransferase (rhodanese) |
| 87 | c1zgxB_ |
|
not modelled |
7.7 |
33 |
PDB header:hydrolase
Chain: B: PDB Molecule:guanyl-specific ribonuclease sa;
PDBTitle: crystal structure of ribonuclease mutant
|
| 88 | d1jl3a_ |
|
not modelled |
7.7 |
11 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 89 | d1i5za2 |
|
not modelled |
7.7 |
15 |
Fold:Double-stranded beta-helix
Superfamily:cAMP-binding domain-like
Family:cAMP-binding domain |
| 90 | d1d2na_ |
|
not modelled |
7.6 |
8 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 91 | d2nxfa1 |
|
not modelled |
7.5 |
9 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:ADPRibase-Mn-like |
| 92 | c2vzaD_ |
|
not modelled |
7.5 |
14 |
PDB header:cell adhesion
Chain: D: PDB Molecule:cell filamentation protein;
PDBTitle: type iv secretion system effector protein bepa
|
| 93 | d2jdig1 |
|
not modelled |
7.4 |
33 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:ATP synthase (F1-ATPase), gamma subunit
Family:ATP synthase (F1-ATPase), gamma subunit |
| 94 | c2zbkB_ |
|
not modelled |
7.4 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:type 2 dna topoisomerase 6 subunit b;
PDBTitle: crystal structure of an intact type ii dna topoisomerase:2 insights into dna transfer mechanisms
|
| 95 | c3j08A_ |
|
not modelled |
7.4 |
18 |
PDB header:hydrolase, metal transport
Chain: A: PDB Molecule:copper-exporting p-type atpase a;
PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa
|
| 96 | c2k23A_ |
|
not modelled |
7.3 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:lipocalin 2;
PDBTitle: solution structure analysis of the rlcn2
|
| 97 | c3gd5D_ |
|
not modelled |
7.3 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from gloeobacter2 violaceus
|
| 98 | d1yp2a2 |
|
not modelled |
7.3 |
15 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
| 99 | c2l3nA_ |
|
not modelled |
7.3 |
13 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna-binding protein rap1, telomere length regulator taz1;
PDBTitle: solution structure of rap1-taz1 fusion protein
|

