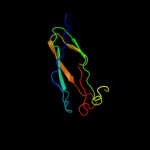
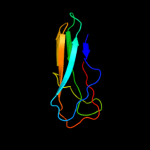
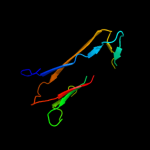
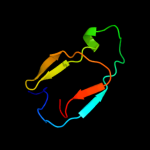
1 c3o0lB_
100.0
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a pfam duf1425 family member (shew_1734) from2 shewanella sp. pv-4 at 1.81 a resolution
2 d2vzsa2
91.2
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain3 c3fn9B_
85.6
11
PDB header: hydrolaseChain: B: PDB Molecule: putative beta-galactosidase;PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis
4 d2co7b1
85.1
11
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PapD-likeFamily: Pilus chaperone5 d3bwuc1
82.8
9
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PapD-likeFamily: Pilus chaperone6 c3isyA_
77.8
9
PDB header: protein bindingChain: A: PDB Molecule: intracellular proteinase inhibitor;PDBTitle: crystal structure of an intracellular proteinase inhibitor (ipi,2 bsu11130) from bacillus subtilis at 2.61 a resolution
7 d1lm8v_
77.5
20
Fold: Prealbumin-likeSuperfamily: VHLFamily: VHL8 c2x41A_
76.2
7
PDB header: hydrolaseChain: A: PDB Molecule: beta-glucosidase;PDBTitle: structure of beta-glucosidase 3b from thermotoga neapolitana2 in complex with glucose
9 c3butA_
71.0
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein af_0446;PDBTitle: crystal structure of protein af_0446 from archaeoglobus fulgidus
10 d1p5va1
62.4
11
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PapD-likeFamily: Pilus chaperone11 c1e9zA_
59.7
9
PDB header: hydrolaseChain: A: PDB Molecule: urease subunit alpha;PDBTitle: crystal structure of helicobacter pylori urease
12 c3rgbA_
59.7
17
PDB header: oxidoreductaseChain: A: PDB Molecule: methane monooxygenase subunit b2;PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
13 c2co7B_
59.4
11
PDB header: fibril proteinChain: B: PDB Molecule: putative fimbriae assembly chaperone;PDBTitle: salmonella enterica safa pilin in complex with the safb2 chaperone (type ii)
14 c1s3rA_
57.7
17
PDB header: toxinChain: A: PDB Molecule: intermedilysin;PDBTitle: crystal structure of the human-specific toxin intermedilysin
15 c3cfuA_
56.7
13
PDB header: lipoproteinChain: A: PDB Molecule: uncharacterized lipoprotein yjha;PDBTitle: crystal structure of the yjha protein from bacillus2 subtilis. northeast structural genomics consortium target3 sr562
16 c1yycA_
54.2
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative late embryogenesis abundant protein;PDBTitle: solution structure of a putative late embryogenesis2 abundant (lea) protein at2g46140.1
17 d2je8a1
53.1
10
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain18 c3ac0B_
52.2
10
PDB header: hydrolaseChain: B: PDB Molecule: beta-glucosidase i;PDBTitle: crystal structure of beta-glucosidase from kluyveromyces marxianus in2 complex with glucose
19 c2kl6A_
51.3
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of the cardb domain of pf1109 from2 pyrococcus furiosus. northeast structural genomics3 consortium target pfr193a
20 c1z9sA_
50.7
11
PDB header: chaperone/immune systemChain: A: PDB Molecule: chaperone protein caf1m;PDBTitle: crystal structure of the native chaperone:subunit:subunit2 caf1m:caf1:caf1 complex
21 c2xfgB_
not modelled
47.7
11
PDB header: hydrolase/sugar binding proteinChain: B: PDB Molecule: endoglucanase 1;PDBTitle: reassembly and co-crystallization of a family 9 processive2 endoglucanase from separately expressed gh9 and cbm3c3 modules
22 c2xbtA_
not modelled
46.1
15
PDB header: sugar binding proteinChain: A: PDB Molecule: cellulosomal scaffoldin;PDBTitle: structure of a scaffoldin carbohydrate-binding module family 3b from2 the cellulosome of bacteroides cellulosolvens: structural diversity3 and implications for carbohydrate binding
23 d2j2za1
not modelled
45.2
10
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PapD-likeFamily: Pilus chaperone24 c3qgaD_
not modelled
43.9
10
PDB header: hydrolaseChain: D: PDB Molecule: fusion of urease beta and gamma subunits;PDBTitle: 3.0 a model of iron containing urease urea2b2 from helicobacter2 mustelae
25 d1pfoa_
not modelled
42.3
19
Fold: PerfringolysinSuperfamily: PerfringolysinFamily: Perfringolysin26 c1pfoA_
not modelled
42.3
19
PDB header: toxinChain: A: PDB Molecule: perfringolysin o;PDBTitle: perfringolysin o
27 d1xo8a_
not modelled
42.2
11
Fold: Immunoglobulin-like beta-sandwichSuperfamily: LEA14-likeFamily: LEA14-like28 c3q48B_
not modelled
41.7
9
PDB header: chaperoneChain: B: PDB Molecule: chaperone cupb2;PDBTitle: crystal structure of pseudomonas aeruginosa cupb2 chaperone
29 d1jz8a2
not modelled
38.3
10
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain30 c3hs0B_
not modelled
33.5
19
PDB header: immune systemChain: B: PDB Molecule: cobra venom factor;PDBTitle: cobra venom factor (cvf) in complex with human factor b
31 c3cu7A_
not modelled
33.0
14
PDB header: immune systemChain: A: PDB Molecule: complement c5;PDBTitle: human complement component 5
32 c2ys4A_
not modelled
32.8
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hydrocephalus-inducing protein homolog;PDBTitle: solution structure of the n-terminal papd-like domain of2 hydin protein from human
33 c3hvnA_
not modelled
32.7
16
PDB header: toxinChain: A: PDB Molecule: hemolysin;PDBTitle: crystal structure of cytotoxin protein suilysin from2 streptococcus suis
34 d1cyga2
not modelled
32.0
17
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Starch-binding domain35 d1qhoa2
not modelled
31.9
17
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Starch-binding domain36 d1yq2a1
not modelled
31.1
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain37 d1i5pa1
not modelled
31.0
24
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: delta-Endotoxin, C-terminal domain38 c1skhA_
not modelled
30.6
26
PDB header: unknown functionChain: A: PDB Molecule: major prion protein 2;PDBTitle: n-terminal (1-30) of bovine prion protein
39 c2c3wB_
not modelled
30.4
22
PDB header: sugar-binding proteinChain: B: PDB Molecule: alpha-amylase g-6;PDBTitle: structure of cbm25 from bacillus halodurans amylase in2 complex with maltotetraose
40 d1l4ia1
not modelled
29.4
11
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PapD-likeFamily: Pilus chaperone41 d1ejxb_
not modelled
28.4
7
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit42 c2f3xA_
not modelled
28.4
9
PDB header: gene regulationChain: A: PDB Molecule: transcription factor fapr;PDBTitle: crystal structure of fapr (in complex with effector)- a2 global regulator of fatty acid biosynthesis in b. subtilis
43 d4ubpb_
not modelled
28.1
4
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit44 d1nbca_
not modelled
27.5
14
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Cellulose-binding domain family III45 c2e6jA_
not modelled
27.5
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hydin protein;PDBTitle: solution structure of the c-terminal papd-like domain from2 human hydin protein
46 c2je8B_
not modelled
25.3
19
PDB header: hydrolaseChain: B: PDB Molecule: beta-mannosidase;PDBTitle: structure of a beta-mannosidase from bacteroides2 thetaiotaomicron
47 c2l8aA_
not modelled
24.3
17
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: structure of a novel cbm3 lacking the calcium-binding site
48 c1l4iA_
not modelled
23.5
10
PDB header: chaperoneChain: A: PDB Molecule: sfae protein;PDBTitle: crystal structure of the periplasmic chaperone sfae
49 d2j5wa3
not modelled
23.1
12
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins50 c2w5fB_
not modelled
23.0
15
PDB header: hydrolaseChain: B: PDB Molecule: endo-1,4-beta-xylanase y;PDBTitle: high resolution crystallographic structure of the2 clostridium thermocellum n-terminal endo-1,4-beta-d-3 xylanase 10b (xyn10b) cbm22-1-gh10 modules complexed with4 xylohexaose
51 d2q0zx2
not modelled
22.8
29
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Sec63 C-terminal domain-like52 c2q0zX_
not modelled
22.4
16
PDB header: protein transportChain: X: PDB Molecule: protein pro2281;PDBTitle: crystal structure of q9p172/sec63 from homo sapiens.2 northeast structural genomics target hr1979.
53 d1e9ya1
not modelled
22.4
10
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit54 c3hftA_
not modelled
19.8
36
PDB header: hydrolaseChain: A: PDB Molecule: wbms, polysaccharide deacetylase involved in o-antigenPDBTitle: crystal structure of a putative polysaccharide deacetylase involved in2 o-antigen biosynthesis (wbms, bb0128) from bordetella bronchiseptica3 at 1.90 a resolution
55 c1qunA_
not modelled
19.2
11
PDB header: chaperone/structural proteinChain: A: PDB Molecule: papd-like chaperone fimc;PDBTitle: x-ray structure of the fimc-fimh chaperone adhesin complex2 from uropathogenic e.coli
56 d1lgta2
not modelled
19.1
29
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases57 d1aoza2
not modelled
19.1
9
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins58 c1sddA_
not modelled
17.0
11
PDB header: blood clottingChain: A: PDB Molecule: coagulation factor v;PDBTitle: crystal structure of bovine factor vai
59 d2je8a3
not modelled
16.4
14
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain60 c3qbtH_
not modelled
15.6
18
PDB header: protein transport/hydrolaseChain: H: PDB Molecule: inositol polyphosphate 5-phosphatase ocrl-1;PDBTitle: crystal structure of ocrl1 540-678 in complex with rab8a:gppnhp
61 d1kula_
not modelled
15.0
16
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Starch-binding domain62 c2x3bB_
not modelled
14.7
22
PDB header: hydrolaseChain: B: PDB Molecule: toxic extracellular endopeptidase;PDBTitle: asap1 inactive mutant e294a, an extracellular toxic zinc2 metalloendopeptidase
63 c2wo4A_
not modelled
13.9
22
PDB header: hydrolaseChain: A: PDB Molecule: glycoside hydrolase, family 9;PDBTitle: 3b' carbohydrate-binding module from the cel9v glycoside2 hydrolase from clostridium thermocellum, in-house data
64 d1nqjb_
not modelled
13.6
11
Fold: CUB-likeSuperfamily: Collagen-binding domainFamily: Collagen-binding domain65 d1f1ua2
not modelled
13.5
20
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases66 c2l0dA_
not modelled
13.1
9
PDB header: cell adhesionChain: A: PDB Molecule: cell surface protein;PDBTitle: solution nmr structure of putative cell surface protein ma_4588 (272-2 376 domain) from methanosarcina acetivorans, northeast structural3 genomics consortium target mvr254a
67 c3zx1A_
not modelled
12.7
16
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, putative;PDBTitle: multicopper oxidase from campylobacter jejuni: a metallo-oxidase
68 d1m1sa_
not modelled
12.7
8
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PapD-likeFamily: MSP-like69 d2vzsa1
not modelled
12.5
9
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain70 d1uura2
not modelled
12.0
6
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: STAT DNA-binding domain71 c1yewI_
not modelled
11.6
16
PDB header: oxidoreductase, membrane proteinChain: I: PDB Molecule: particulate methane monooxygenase, b subunit;PDBTitle: crystal structure of particulate methane monooxygenase
72 d1f1xa2
not modelled
11.6
13
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases73 d1ex0a2
not modelled
11.5
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Transglutaminase, two C-terminal domainsFamily: Transglutaminase, two C-terminal domains74 d2je8a2
not modelled
11.4
0
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain75 d1jzga_
not modelled
11.1
3
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like76 d1e42a1
not modelled
11.0
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Clathrin adaptor appendage domainFamily: Alpha-adaptin ear subdomain-like77 d2k5qa1
not modelled
10.7
23
Fold: OB-foldSuperfamily: BC4932-likeFamily: BC4932-like78 d1ifra_
not modelled
10.6
14
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Lamin A/C globular tail domainFamily: Lamin A/C globular tail domain79 d1nwpa_
not modelled
10.4
12
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like80 d1mpya1
not modelled
10.3
14
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases81 c3gekA_
not modelled
10.2
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative thioesterase yhda;PDBTitle: crystal structure of putative thioesterase yhda from lactococcus2 lactis. northeast structural genomics consortium target kr113
82 c2lllA_
not modelled
10.1
11
PDB header: structural proteinChain: A: PDB Molecule: lamin-b2;PDBTitle: solution nmr structure of c-terminal globular domain of human lamin-2 b2, northeast structural genomics consortium target hr8546a
83 c2h47C_
not modelled
9.9
3
PDB header: oxidoreductase/electron transportChain: C: PDB Molecule: azurin;PDBTitle: crystal structure of an electron transfer complex between2 aromatic amine dephydrogenase and azurin from alcaligenes3 faecalis (form 1)
84 d1h6ya_
not modelled
9.4
11
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: CBM2285 d1azca_
not modelled
9.4
3
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like86 c2r39A_
not modelled
9.3
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: fixg-related protein;PDBTitle: crystal structure of fixg-related protein from vibrio parahaemolyticus
87 d2j5wa1
not modelled
9.2
15
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins88 c2opkC_
not modelled
9.2
23
PDB header: isomeraseChain: C: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a putative mannose-6-phosphate isomerase2 (reut_a1446) from ralstonia eutropha jmp134 at 2.10 a resolution
89 c1qpxA_
not modelled
9.2
10
PDB header: chaperoneChain: A: PDB Molecule: papd chaperone;PDBTitle: crystal structures of self-capping papd chaperone homodimers
90 d3eipa_
not modelled
8.9
13
Fold: FKBP-likeSuperfamily: Colicin E3 immunity proteinFamily: Colicin E3 immunity protein91 d1q38a_
not modelled
8.9
25
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Fibronectin type IIIFamily: Fibronectin type III92 c2h7fX_
not modelled
8.9
9
PDB header: isomerase/dnaChain: X: PDB Molecule: dna topoisomerase 1;PDBTitle: structure of variola topoisomerase covalently bound to dna
93 d1f53a_
not modelled
8.7
33
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Killer toxin-like protein SKLP94 d1cc3a_
not modelled
8.6
7
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like95 c3qhtD_
not modelled
8.6
21
PDB header: de novo proteinChain: D: PDB Molecule: monobody ysmb-1;PDBTitle: crystal structure of the monobody ysmb-1 bound to yeast sumo
96 c1cyxA_
not modelled
8.6
26
PDB header: electron transportChain: A: PDB Molecule: cyoa;PDBTitle: quinol oxidase (periplasmic fragment of subunit ii with2 engineered cu-a binding site)(cyoa)
97 d1cyxa_
not modelled
8.6
26
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Periplasmic domain of cytochrome c oxidase subunit II98 d2ccwa1
not modelled
8.5
4
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like99 c2rk0B_
not modelled
8.3
14
PDB header: oxidoreductaseChain: B: PDB Molecule: glyoxalase/bleomycin resistance protein/dioxygenase domain;PDBTitle: crystal structure of glyoxylase/bleomycin resistance2 protein/dioxygenase domain from frankia sp. ean1pec