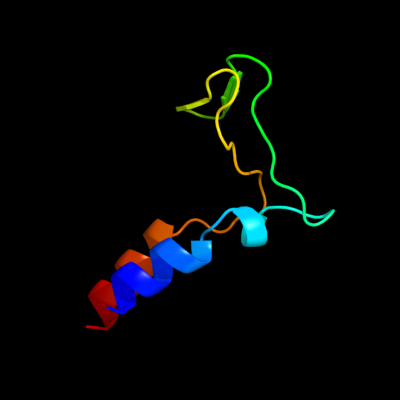
1 c2rfpA_
75.8
24
PDB header: hydrolaseChain: A: PDB Molecule: putative ntp pyrophosphohydrolase;PDBTitle: crystal structure of putative ntp pyrophosphohydrolase2 (yp_001813558.1) from exiguobacterium sibiricum 255-15 at 1.74 a3 resolution
2 c1vgmB_
46.4
16
PDB header: transferaseChain: B: PDB Molecule: 378aa long hypothetical citrate synthase;PDBTitle: crystal structure of an isozyme of citrate synthase from sulfolbus2 tokodaii strain7
3 d1hw1a1
44.4
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: GntR-like transcriptional regulators4 c2r26C_
37.1
13
PDB header: transferaseChain: C: PDB Molecule: citrate synthase;PDBTitle: the structure of the ternary complex of carboxymethyl2 coenzyme a and oxalateacetate with citrate synthase from3 the thermophilic archaeonthermoplasma acidophilum
5 c3ic7A_
36.8
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator of gntr family2 from bacteroides thetaiotaomicron
6 c2wv0H_
34.4
21
PDB header: transcriptionChain: H: PDB Molecule: hth-type transcriptional repressor yvoa;PDBTitle: crystal structure of the gntr-hutc family member yvoa from2 bacillus subtilis
7 d1o7xa_
34.0
11
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase8 c1e2xA_
33.4
25
PDB header: transcriptional regulationChain: A: PDB Molecule: fatty acid metabolism regulator protein;PDBTitle: fadr, fatty acid responsive transcription factor from e.2 coli
9 c3neuA_
30.0
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin1836 protein;PDBTitle: the crystal structure of a functionally-unknown protein lin1836 from2 listeria innocua clip11262
10 d1v4ra1
28.7
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: GntR-like transcriptional regulators11 c2vcbA_
28.4
30
PDB header: hydrolaseChain: A: PDB Molecule: alpha-n-acetylglucosaminidase;PDBTitle: family 89 glycoside hydrolase from clostridium perfringens2 in complex with pugnac
12 d1ioma_
27.7
15
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase13 c3by6C_
26.4
12
PDB header: transcription regulatorChain: C: PDB Molecule: predicted transcriptional regulator;PDBTitle: crystal structure of a transcriptional regulator from oenococcus oeni
14 d1k3pa_
25.2
19
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase15 c3eetA_
24.6
9
PDB header: transcription regulatorChain: A: PDB Molecule: putative gntr-family transcriptional regulator;PDBTitle: crystal structure of putative gntr-family transcriptional2 regulator
16 c3bwgA_
22.8
13
PDB header: transcription regulatorChain: A: PDB Molecule: uncharacterized hth-type transcriptional regulator yydk;PDBTitle: the crystal structure of possible transcriptional regulator yydk from2 bacillus subtilis subsp. subtilis str. 168
17 c2p2wA_
22.7
14
PDB header: transferaseChain: A: PDB Molecule: citrate synthase;PDBTitle: crystal structure of citrate synthase from thermotoga maritima msb8
18 c3edpB_
22.6
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: lin2111 protein;PDBTitle: the crystal structure of the protein lin2111 (functionally unknown)2 from listeria innocua clip11262
19 c3tqnC_
22.1
20
PDB header: transcriptionChain: C: PDB Molecule: transcriptional regulator, gntr family;PDBTitle: structure of the transcriptional regulator of the gntr family, from2 coxiella burnetii.
20 d1a59a_
22.0
18
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase21 d1r9la_
not modelled
21.3
18
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like22 c3tqgA_
not modelled
20.9
17
PDB header: transferaseChain: A: PDB Molecule: 2-methylcitrate synthase;PDBTitle: structure of the 2-methylcitrate synthase (prpc) from coxiella2 burnetii
23 c3o8jH_
not modelled
20.6
19
PDB header: transferaseChain: H: PDB Molecule: 2-methylcitrate synthase;PDBTitle: crystal structure of 2-methylcitrate synthase (prpc) from salmonella2 typhimurium
24 c2du9A_
not modelled
18.9
25
PDB header: transcriptionChain: A: PDB Molecule: predicted transcriptional regulators;PDBTitle: crystal structure of the transcriptional factor from c.glutamicum
25 c3fmsA_
not modelled
18.5
17
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, gntr family;PDBTitle: crystal structure of tm0439, a gntr transcriptional2 regulator
26 c2h12C_
not modelled
17.6
19
PDB header: transferaseChain: C: PDB Molecule: citrate synthase;PDBTitle: structure of acetobacter aceti citrate synthase complexed2 with oxaloacetate and carboxymethyldethia coenzyme a (cmx)
27 d1csca_
not modelled
15.9
13
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase28 d1hc1a2
not modelled
15.8
21
Fold: Di-copper centre-containing domainSuperfamily: Di-copper centre-containing domainFamily: Hemocyanin middle domain29 d2fzta1
not modelled
15.7
41
Fold: Methionine synthase domain-likeSuperfamily: TM0693-likeFamily: TM0693-like30 c3hhsA_
not modelled
15.6
35
PDB header: oxidoreductaseChain: A: PDB Molecule: phenoloxidase subunit 2;PDBTitle: crystal structure of manduca sexta prophenoloxidase
31 d3bwga1
not modelled
15.5
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: GntR-like transcriptional regulators32 d1llaa2
not modelled
13.9
15
Fold: Di-copper centre-containing domainSuperfamily: Di-copper centre-containing domainFamily: Hemocyanin middle domain33 d1bh9b_
not modelled
13.1
27
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs34 c2i9nA_
not modelled
12.8
70
PDB header: de novo proteinChain: A: PDB Molecule: mhb4a peptide;PDBTitle: design of bivalent miniprotein consisting of two2 independent elements, a b-hairpin peptide and a-helix3 peptide, tethered by four glycines
35 c2i9oA_
not modelled
12.5
70
PDB header: de novo proteinChain: A: PDB Molecule: mhb8a peptide;PDBTitle: design of bivalent miniprotein consisting of two2 independent elements, a b-hairpin peptide and a-helix3 peptide, tethered by eight glycines
36 d1e8ca2
not modelled
11.9
21
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain37 c3fkeB_
not modelled
11.5
40
PDB header: rna binding proteinChain: B: PDB Molecule: polymerase cofactor vp35;PDBTitle: structure of the ebola vp35 interferon inhibitory domain
38 c2ibpB_
not modelled
11.4
15
PDB header: transferaseChain: B: PDB Molecule: citrate synthase;PDBTitle: crystal structure of citrate synthase from pyrobaculum aerophilum
39 d2hs5a1
not modelled
11.2
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: GntR-like transcriptional regulators40 c1iq8B_
not modelled
11.1
26
PDB header: transferaseChain: B: PDB Molecule: archaeosine trna-guanine transglycosylase;PDBTitle: crystal structure of archaeosine trna-guanine2 transglycosylase from pyrococcus horikoshii
41 d1vi0a2
not modelled
10.4
18
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain42 c3ks8D_
not modelled
9.2
40
PDB header: viral protein/rnaChain: D: PDB Molecule: polymerase cofactor vp35;PDBTitle: crystal structure of reston ebolavirus vp35 rna binding2 domain in complex with 18bp dsrna
43 d2g7sa2
not modelled
8.4
20
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain44 c3f8mA_
not modelled
8.4
18
PDB header: transcriptionChain: A: PDB Molecule: gntr-family protein transcriptional regulator;PDBTitle: crystal structure of phnf from mycobacterium smegmatis
45 d1rh7a_
not modelled
8.3
18
Fold: ResistinSuperfamily: ResistinFamily: Resistin46 d1aj8a_
not modelled
8.0
18
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase47 d1nb5i_
not modelled
7.9
40
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Cystatins48 d1o98a1
not modelled
7.8
40
Fold: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domainSuperfamily: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domainFamily: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain49 c1nolA_
not modelled
7.5
18
PDB header: oxygen transportChain: A: PDB Molecule: hemocyanin (subunit type ii);PDBTitle: oxygenated hemocyanin (subunit type ii)
50 d1atga_
not modelled
7.4
29
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like51 c3mvnA_
not modelled
7.3
19
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate:l-alanyl-gamma-d-glutamayl-medo-PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamayl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
52 d2i10a2
not modelled
7.2
20
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain53 d1bvsa2
not modelled
7.1
21
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: DNA helicase RuvA subunit, middle domain54 c1hcyE_
not modelled
7.0
25
PDB header: oxygen transportChain: E: PDB Molecule: arthropodan hemocyanin;PDBTitle: crystal structure of hexameric haemocyanin from panulirus interruptus2 refined at 3.2 angstroms resolution
55 d1fi4a2
not modelled
6.7
10
Fold: Ferredoxin-likeSuperfamily: GHMP Kinase, C-terminal domainFamily: Mevalonate 5-diphosphate decarboxylase56 c3ajvA_
not modelled
6.5
14
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: splicing endonuclease from aeropyrum pernix
57 d1z1za1
not modelled
6.5
22
Fold: Phage tail protein-likeSuperfamily: Phage tail protein-likeFamily: Lambda phage gpU-like58 d1dpya_
not modelled
6.4
38
Fold: Phospholipase A2, PLA2Superfamily: Phospholipase A2, PLA2Family: Vertebrate phospholipase A259 d1g8fa3
not modelled
6.4
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ATP sulfurylase C-terminal domain60 c3gwjE_
not modelled
6.3
33
PDB header: oxygen transportChain: E: PDB Molecule: arylphorin;PDBTitle: crystal structure of antheraea pernyi arylphorin
61 d1yj5a2
not modelled
6.0
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases62 d1p3da2
not modelled
5.9
26
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain63 c1wsuA_
not modelled
5.9
15
PDB header: translation/rnaChain: A: PDB Molecule: selenocysteine-specific elongation factor;PDBTitle: c-terminal domain of elongation factor selb complexed with2 secis rna
64 d2zdra2
not modelled
5.8
21
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: NeuB-like65 c1yrlD_
not modelled
5.7
9
PDB header: oxidoreductaseChain: D: PDB Molecule: ketol-acid reductoisomerase;PDBTitle: escherichia coli ketol-acid reductoisomerase
66 c2xkyI_
not modelled
5.6
30
PDB header: metal transportChain: I: PDB Molecule: inward rectifier potassium channel 2;PDBTitle: single particle analysis of kir2.1nc_4 in negative stain
67 d1k6ma1
not modelled
5.6
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, kinase domain68 d2o97b1
not modelled
5.5
27
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein69 c2ov2O_
not modelled
5.5
22
PDB header: protein binding/transferaseChain: O: PDB Molecule: serine/threonine-protein kinase pak 4;PDBTitle: the crystal structure of the human rac3 in complex with the crib2 domain of human p21-activated kinase 4 (pak4)
70 d1bifa1
not modelled
5.4
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, kinase domain71 c2odbB_
not modelled
5.3
33
PDB header: protein bindingChain: B: PDB Molecule: serine/threonine-protein kinase pak 6;PDBTitle: the crystal structure of human cdc42 in complex with the crib domain2 of human p21-activated kinase 6 (pak6)