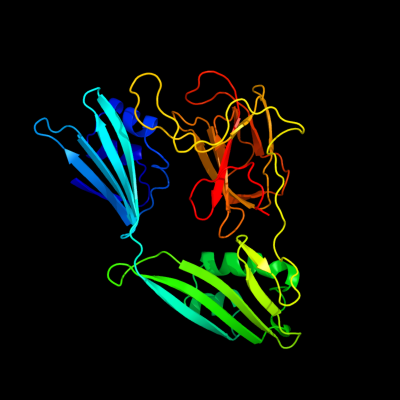
1 c2gu1A_
100.0
42
PDB header: hydrolaseChain: A: PDB Molecule: zinc peptidase;PDBTitle: crystal structure of a zinc containing peptidase from2 vibrio cholerae
2 c2hsiB_
100.0
37
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative peptidase m23;PDBTitle: crystal structure of putative peptidase m23 from2 pseudomonas aeruginosa, new york structural genomics3 consortium
3 c3nyyA_
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: putative glycyl-glycine endopeptidase lytm;PDBTitle: crystal structure of a putative glycyl-glycine endopeptidase lytm2 (rumgna_02482) from ruminococcus gnavus atcc 29149 at 1.60 a3 resolution
4 d1qwya_
100.0
33
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Peptidoglycan hydrolase LytM5 c2b44A_
100.0
38
PDB header: hydrolaseChain: A: PDB Molecule: glycyl-glycine endopeptidase lytm;PDBTitle: truncated s. aureus lytm, p 32 2 1 crystal form
6 c3it5B_
100.0
21
PDB header: hydrolaseChain: B: PDB Molecule: protease lasa;PDBTitle: crystal structure of the lasa virulence factor from pseudomonas2 aeruginosa
7 c3csqC_
99.9
23
PDB header: hydrolaseChain: C: PDB Molecule: morphogenesis protein 1;PDBTitle: crystal and cryoem structural studies of a cell wall2 degrading enzyme in the bacteriophage phi29 tail
8 d2f3ga_
97.1
20
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like9 d1glaf_
97.0
19
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like10 d2gpra_
96.7
17
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like11 d1gpra_
96.7
15
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like12 c2aukA_
95.8
16
PDB header: transferaseChain: A: PDB Molecule: dna-directed rna polymerase beta' chain;PDBTitle: structure of e. coli rna polymerase beta' g/g' insert
13 d1brwa3
93.8
24
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain14 d1dcza_
93.3
20
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains15 d1ci3m2
93.3
18
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: Cytochrome f, small domain16 d1e2wa2
93.2
19
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: Cytochrome f, small domain17 d2tpta3
92.5
20
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain18 c2kccA_
92.2
21
PDB header: ligaseChain: A: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: solution structure of biotinoyl domain from human acetyl-2 coa carboxylase 2
19 c2ejgD_
91.9
21
PDB header: ligaseChain: D: PDB Molecule: 149aa long hypothetical methylmalonyl-coa decarboxylasePDBTitle: crystal structure of the biotin protein ligase (mutation r48a) and2 biotin carboxyl carrier protein complex from pyrococcus horikoshii3 ot3
20 c1otpA_
91.6
20
PDB header: phosphorylaseChain: A: PDB Molecule: thymidine phosphorylase;PDBTitle: structural and theoretical studies suggest domain movement produces an2 active conformation of thymidine phosphorylase
21 c2j0fC_
not modelled
91.6
25
PDB header: transferaseChain: C: PDB Molecule: thymidine phosphorylase;PDBTitle: structural basis for non-competitive product inhibition in2 human thymidine phosphorylase: implication for drug design
22 c1e2vB_
not modelled
91.4
17
PDB header: electron transport proteinsChain: B: PDB Molecule: cytochrome f;PDBTitle: n153q mutant of cytochrome f from chlamydomonas reinhardtii
23 c1ctmA_
not modelled
91.1
17
PDB header: electron transport(cytochrome)Chain: A: PDB Molecule: cytochrome f;PDBTitle: crystal structure of chloroplast cytochrome f reveals a2 novel cytochrome fold and unexpected heme ligation
24 c1q90A_
not modelled
90.7
17
PDB header: photosynthesisChain: A: PDB Molecule: apocytochrome f;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
25 c3h5qA_
not modelled
90.6
20
PDB header: transferaseChain: A: PDB Molecule: pyrimidine-nucleoside phosphorylase;PDBTitle: crystal structure of a putative pyrimidine-nucleoside phosphorylase2 from staphylococcus aureus
26 c2jxmB_
not modelled
90.2
16
PDB header: electron transportChain: B: PDB Molecule: cytochrome f;PDBTitle: ensemble of twenty structures of the prochlorothrix2 hollandica plastocyanin- cytochrome f complex
27 d1bdoa_
not modelled
90.0
18
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains28 c2b8gA_
not modelled
90.0
17
PDB header: biosynthetic proteinChain: A: PDB Molecule: biotin/lipoyl attachment protein;PDBTitle: solution structure of bacillus subtilis blap biotinylated-2 form (energy minimized mean structure)
29 d1uoua3
not modelled
89.9
22
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain30 c3n6rK_
not modelled
89.5
25
PDB header: ligaseChain: K: PDB Molecule: propionyl-coa carboxylase, alpha subunit;PDBTitle: crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
31 c2dsjA_
not modelled
88.2
37
PDB header: transferaseChain: A: PDB Molecule: pyrimidine-nucleoside (thymidine) phosphorylase;PDBTitle: crystal structure of project id tt0128 from thermus thermophilus hb8
32 c1tu2B_
not modelled
87.2
16
PDB header: electron transportChain: B: PDB Molecule: apocytochrome f;PDBTitle: the complex of nostoc cytochrome f and plastocyanin determin with2 paramagnetic nmr. based on the structures of cytochrome f and3 plastocyanin, 10 structures
33 d1ghja_
not modelled
87.2
19
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains34 c3lnnB_
not modelled
87.1
22
PDB header: metal transportChain: B: PDB Molecule: membrane fusion protein (mfp) heavy metal cation effluxPDBTitle: crystal structure of zneb from cupriavidus metallidurans
35 c1t5eB_
not modelled
86.8
30
PDB header: transport proteinChain: B: PDB Molecule: multidrug resistance protein mexa;PDBTitle: the structure of mexa
36 c2k33A_
not modelled
86.2
30
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: acra;PDBTitle: solution structure of an n-glycosylated protein using in2 vitro glycosylation
37 c2f1mA_
not modelled
86.1
17
PDB header: transport proteinChain: A: PDB Molecule: acriflavine resistance protein a;PDBTitle: conformational flexibility in the multidrug efflux system protein acra
38 c1brwB_
not modelled
85.7
22
PDB header: transferaseChain: B: PDB Molecule: protein (pyrimidine nucleoside phosphorylase);PDBTitle: the crystal structure of pyrimidine nucleoside2 phosphorylase in a closed conformation
39 d1o78a_
not modelled
85.2
39
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains40 c3fppB_
not modelled
84.7
26
PDB header: membrane proteinChain: B: PDB Molecule: macrolide-specific efflux protein maca;PDBTitle: crystal structure of e.coli maca
41 d1vf7a_
not modelled
84.7
27
Fold: HlyD-like secretion proteinsSuperfamily: HlyD-like secretion proteinsFamily: HlyD-like secretion proteins42 d1iyua_
not modelled
84.2
24
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains43 c2aujD_
not modelled
83.4
20
PDB header: transferaseChain: D: PDB Molecule: dna-directed rna polymerase beta' chain;PDBTitle: structure of thermus aquaticus rna polymerase beta'-subunit2 insert
44 d1qjoa_
not modelled
83.4
24
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains45 c2ejmA_
not modelled
82.5
13
PDB header: ligaseChain: A: PDB Molecule: methylcrotonoyl-coa carboxylase subunit alpha;PDBTitle: solution structure of ruh-072, an apo-biotnyl domain form2 human acetyl coenzyme a carboxylase
46 c3camB_
not modelled
82.4
23
PDB header: gene regulationChain: B: PDB Molecule: cold-shock domain family protein;PDBTitle: crystal structure of the cold shock domain protein from neisseria2 meningitidis
47 c2q8iB_
not modelled
81.0
26
PDB header: transferaseChain: B: PDB Molecule: dihydrolipoyllysine-residue acetyltransferase component ofPDBTitle: pyruvate dehydrogenase kinase isoform 3 in complex with antitumor drug2 radicicol
48 d1tu2b2
not modelled
80.8
23
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: Cytochrome f, small domain49 c2qf7A_
not modelled
80.3
21
PDB header: ligaseChain: A: PDB Molecule: pyruvate carboxylase protein;PDBTitle: crystal structure of a complete multifunctional pyruvate carboxylase2 from rhizobium etli
50 c2dn8A_
not modelled
80.1
22
PDB header: ligaseChain: A: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: solution structure of rsgi ruh-053, an apo-biotin carboxy2 carrier protein from human transcarboxylase
51 c2l5tA_
not modelled
79.6
17
PDB header: transferaseChain: A: PDB Molecule: lipoamide acyltransferase;PDBTitle: solution nmr structure of e2 lipoyl domain from thermoplasma2 acidophilum
52 d1y8ob1
not modelled
79.2
26
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains53 d1laba_
not modelled
78.5
17
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains54 d1gjxa_
not modelled
77.8
16
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains55 c2e75C_
not modelled
75.6
17
PDB header: photosynthesisChain: C: PDB Molecule: apocytochrome f;PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
56 d1k8ma_
not modelled
75.3
13
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains57 c3fmcC_
not modelled
75.3
20
PDB header: hydrolaseChain: C: PDB Molecule: putative succinylglutamate desuccinylase / aspartoacylase;PDBTitle: crystal structure of a putative succinylglutamate desuccinylase /2 aspartoacylase family protein (sama_0604) from shewanella amazonensis3 sb2b at 1.80 a resolution
58 d2pnrc1
not modelled
74.7
26
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains59 c2qj8B_
not modelled
74.2
8
PDB header: hydrolaseChain: B: PDB Molecule: mlr6093 protein;PDBTitle: crystal structure of an aspartoacylase family protein (mlr6093) from2 mesorhizobium loti maff303099 at 2.00 a resolution
60 c2dneA_
not modelled
73.8
30
PDB header: transferaseChain: A: PDB Molecule: dihydrolipoyllysine-residue acetyltransferasePDBTitle: solution structure of rsgi ruh-058, a lipoyl domain of2 human 2-oxoacid dehydrogenase
61 c2dncA_
not modelled
73.4
30
PDB header: transferaseChain: A: PDB Molecule: pyruvate dehydrogenase protein x component;PDBTitle: solution structure of rsgi ruh-054, a lipoyl domain from2 human 2-oxoacid dehydrogenase
62 c3h9iB_
not modelled
72.5
22
PDB header: transport proteinChain: B: PDB Molecule: cation efflux system protein cusb;PDBTitle: crystal structure of the membrane fusion protein cusb from escherichia2 coli
63 d1h95a_
not modelled
72.5
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like64 c3aqqD_
not modelled
71.9
20
PDB header: dna binding proteinChain: D: PDB Molecule: calcium-regulated heat stable protein 1;PDBTitle: crystal structure of human crhsp-24
65 c2vbcA_
not modelled
69.2
19
PDB header: hydrolaseChain: A: PDB Molecule: dengue 4 ns3 full-length protein;PDBTitle: crystal structure of the ns3 protease-helicase from dengue2 virus
66 c3a0jB_
not modelled
68.6
18
PDB header: transcriptionChain: B: PDB Molecule: cold shock protein;PDBTitle: crystal structure of cold shock protein 1 from thermus2 thermophilus hb8
67 c2jkuA_
not modelled
67.0
37
PDB header: ligaseChain: A: PDB Molecule: propionyl-coa carboxylase alpha chain,PDBTitle: crystal structure of the n-terminal region of the biotin2 acceptor domain of human propionyl-coa carboxylase
68 d2es2a1
not modelled
66.9
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like69 d1pmra_
not modelled
66.3
22
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains70 c3na6A_
not modelled
62.2
27
PDB header: hydrolaseChain: A: PDB Molecule: succinylglutamate desuccinylase/aspartoacylase;PDBTitle: crystal structure of a succinylglutamate desuccinylase (tm1040_2694)2 from silicibacter sp. tm1040 at 2.00 a resolution
71 d1mjca_
not modelled
59.9
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like72 d1c9oa_
not modelled
58.8
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like73 c2ytyA_
not modelled
57.7
24
PDB header: rna binding proteinChain: A: PDB Molecule: cold shock domain-containing protein e1;PDBTitle: solution structure of the fourth cold-shock domain of the human2 kiaa0885 protein (unr protein)
74 c3cdxB_
not modelled
56.7
15
PDB header: hydrolaseChain: B: PDB Molecule: succinylglutamatedesuccinylase/aspartoacylase;PDBTitle: crystal structure of2 succinylglutamatedesuccinylase/aspartoacylase from3 rhodobacter sphaeroides
75 d1g6pa_
not modelled
56.4
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like76 c2bh8B_
not modelled
56.2
18
PDB header: transcriptionChain: B: PDB Molecule: 1b11;PDBTitle: combinatorial protein 1b11
77 c3ozxA_
not modelled
56.1
15
PDB header: hydrolase, translationChain: A: PDB Molecule: rnase l inhibitor;PDBTitle: crystal structure of abce1 of sulfolubus solfataricus (-fes domain)
78 c2k5nA_
not modelled
56.1
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative cold-shock protein;PDBTitle: solution nmr structure of the n-terminal domain of protein2 eca1580 from erwinia carotovora, northeast structural3 genomics consortium target ewr156a
79 c2q6oB_
not modelled
54.7
31
PDB header: biosynthetic proteinChain: B: PDB Molecule: hypothetical protein;PDBTitle: sall-y70t with sam and cl
80 c2zbvC_
not modelled
54.6
22
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized conserved protein;PDBTitle: crystal structure of uncharacterized conserved protein from thermotoga2 maritima
81 d1o4ua2
not modelled
53.7
21
Fold: alpha/beta-HammerheadSuperfamily: Nicotinate/Quinolinate PRTase N-terminal domain-likeFamily: NadC N-terminal domain-like82 c2xoaA_
not modelled
52.8
34
PDB header: metal transportChain: A: PDB Molecule: ryanodine receptor 1;PDBTitle: crystal structure of the n-terminal three domains of the2 skeletal muscle ryanodine receptor (ryr1)
83 c1rqrA_
not modelled
51.8
16
PDB header: transferaseChain: A: PDB Molecule: 5'-fluoro-5'-deoxyadenosine synthase;PDBTitle: crystal structure and mechanism of a bacterial fluorinating enzyme,2 product complex
84 c1xjvA_
not modelled
51.1
12
PDB header: transcription/dnaChain: A: PDB Molecule: protection of telomeres 1;PDBTitle: crystal structure of human pot1 bound to telomeric single-2 stranded dna (ttagggttag)
85 d1qapa2
not modelled
48.3
11
Fold: alpha/beta-HammerheadSuperfamily: Nicotinate/Quinolinate PRTase N-terminal domain-likeFamily: NadC N-terminal domain-like86 c3ilaG_
not modelled
48.0
34
PDB header: signaling proteinChain: G: PDB Molecule: ryanodine receptor 1;PDBTitle: crystal structure of rabbit ryanodine receptor 1 n-terminal domain (9-2 205)
87 c3n9tA_
not modelled
46.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: pnpc;PDBTitle: cryatal structure of hydroxyquinol 1,2-dioxygenase from pseudomonas2 putida dll-e4
88 c3trzE_
not modelled
46.0
14
PDB header: rna binding protein/rnaChain: E: PDB Molecule: protein lin-28 homolog a;PDBTitle: mouse lin28a in complex with let-7d microrna pre-element
89 d1e0ga_
not modelled
45.9
31
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain90 c2l9yA_
not modelled
45.7
24
PDB header: sugar binding proteinChain: A: PDB Molecule: cvnh-lysm lectin;PDBTitle: solution structure of the mocvnh-lysm module from the rice blast2 fungus magnaporthe oryzae protein (mgg_03307)
91 c1wu8B_
not modelled
44.6
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein ph0463;PDBTitle: crystal structure of project ph0463 from pyrococcus horikoshii ot3
92 c1wydB_
not modelled
44.5
15
PDB header: ligaseChain: B: PDB Molecule: hypothetical aspartyl-trna synthetase;PDBTitle: crystal structure of aspartyl-trna synthetase from sulfolobus tokodaii
93 d2ns0a1
not modelled
43.5
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like94 d1qpoa2
not modelled
41.5
21
Fold: alpha/beta-HammerheadSuperfamily: Nicotinate/Quinolinate PRTase N-terminal domain-likeFamily: NadC N-terminal domain-like95 c3a5dB_
not modelled
41.5
20
PDB header: hydrolaseChain: B: PDB Molecule: v-type atp synthase alpha chain;PDBTitle: inter-subunit interaction and quaternary rearrangement2 defined by the central stalk of prokaryotic v1-atpase
96 d1mzya2
not modelled
41.3
30
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins97 c1fpyE_
not modelled
40.0
35
PDB header: ligaseChain: E: PDB Molecule: glutamine synthetase;PDBTitle: crystal structure of glutamine synthetase from salmonella2 typhimurium with inhibitor phosphinothricin
98 c1b8aB_
not modelled
39.9
22
PDB header: ligaseChain: B: PDB Molecule: protein (aspartyl-trna synthetase);PDBTitle: aspartyl-trna synthetase
99 c1htoB_
not modelled
39.2
35
PDB header: ligaseChain: B: PDB Molecule: glutamine synthetase;PDBTitle: crystallographic structure of a relaxed glutamine synthetase from2 mycobacterium tuberculosis
100 d1hcza2
not modelled
39.0
13
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: Cytochrome f, small domain101 c2djpA_
not modelled
37.5
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein sb145;PDBTitle: the solution structure of the lysm domain of human2 hypothetical protein sb145
102 c3imiB_
not modelled
37.5
10
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hit family protein;PDBTitle: 2.01 angstrom resolution crystal structure of a hit family protein2 from bacillus anthracis str. 'ames ancestor'
103 c2ywfA_
not modelled
36.7
19
PDB header: translationChain: A: PDB Molecule: gtp-binding protein lepa;PDBTitle: crystal structure of gmppnp-bound lepa from aquifex aeolicus
104 d2ijob1
not modelled
35.7
31
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases105 c3n7xA_
not modelled
35.3
23
PDB header: virusChain: A: PDB Molecule: capsid protein;PDBTitle: crystal structure of penaeus stylirostris densovirus capsid
106 c3qr5B_
not modelled
35.3
27
PDB header: signaling proteinChain: B: PDB Molecule: cardiac ca2+ release channel;PDBTitle: structure of the first domain of a cardiac ryanodine receptor mutant2 with exon 3 deleted
107 c3o0mB_
not modelled
35.1
37
PDB header: hydrolaseChain: B: PDB Molecule: hit family protein;PDBTitle: crystal structure of a zn-bound histidine triad family protein from2 mycobacterium smegmatis
108 d1f52a2
not modelled
35.0
30
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: Glutamine synthetase catalytic domain109 d1rqpa1
not modelled
33.9
14
Fold: Bacterial fluorinating enzyme, C-terminal domainSuperfamily: Bacterial fluorinating enzyme, C-terminal domainFamily: Bacterial fluorinating enzyme, C-terminal domain110 d1uwfa1
not modelled
33.9
23
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits111 d2awna2
not modelled
33.8
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like112 c2kcmA_
not modelled
33.8
18
PDB header: nucleic acid binding proteinChain: A: PDB Molecule: cold shock domain family protein;PDBTitle: solution nmr structure of the n-terminal ob-domain of so_1732 from2 shewanella oneidensis. northeast structural genomics consortium3 target sor210a.
113 d1k82a2
not modelled
32.8
14
Fold: N-terminal domain of MutM-like DNA repair proteinsSuperfamily: N-terminal domain of MutM-like DNA repair proteinsFamily: N-terminal domain of MutM-like DNA repair proteins114 c2ghiD_
not modelled
32.4
34
PDB header: transport proteinChain: D: PDB Molecule: transport protein;PDBTitle: crystal structure of plasmodium yoelii multidrug resistance2 protein 2
115 c2xhaB_
not modelled
32.0
26
PDB header: transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of domain 2 of thermotoga maritima n-utilization2 substance g (nusg)
116 d3pccm_
not modelled
31.8
20
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase117 c1x65A_
not modelled
31.7
22
PDB header: rna binding proteinChain: A: PDB Molecule: unr protein;PDBTitle: solution structure of the third cold-shock domain of the human2 kiaa0885 protein (unr protein)
118 c3e3xA_
not modelled
31.6
15
PDB header: hydrolaseChain: A: PDB Molecule: bipa;PDBTitle: the c-terminal part of bipa protein from vibrio parahaemolyticus rimd2 2210633
119 c2quoA_
not modelled
31.1
17
PDB header: toxinChain: A: PDB Molecule: heat-labile enterotoxin b chain;PDBTitle: crystal structure of c terminal fragment of clostridium2 perfringens enterotoxin
120 c2j9iL_
not modelled
30.7
20
PDB header: ligaseChain: L: PDB Molecule: glutamate-ammonia ligase domain-containingPDBTitle: lengsin is a survivor of an ancient family of class i2 glutamine synthetases in eukaryotes that has undergone3 evolutionary re-engineering for a tissue-specific role4 in the vertebrate eye lens.