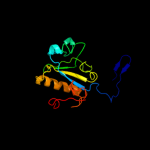
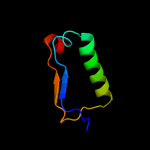
1 c1s2jA_
100.0
43
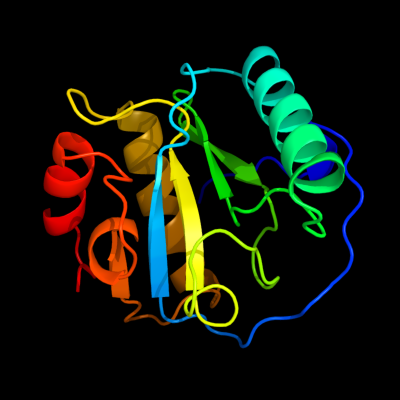
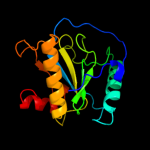
PDB header: hydrolaseChain: A: PDB Molecule: peptidoglycan recognition protein sa cg11709-pa;PDBTitle: crystal structure of the drosophila pattern-recognition2 receptor pgrp-sa
2 d1sxra_
100.0
43
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like3 d2f2la1
100.0
32
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like4 d2cb3a1
100.0
44
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like5 d1ycka1
100.0
45
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like6 c2rkqA_
100.0
40
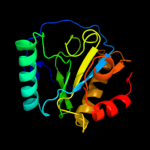
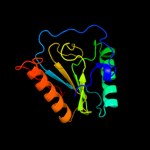
PDB header: immune systemChain: A: PDB Molecule: peptidoglycan-recognition protein-sd;PDBTitle: crystal structure of drosophila peptidoglycan recognition2 protein sd (pgrp-sd)
7 d2f2lx1
100.0
45
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like8 c2xz4A_
100.0
37
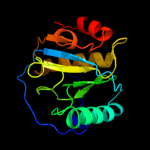
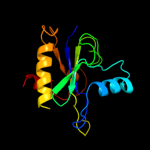
PDB header: immune systemChain: A: PDB Molecule: peptidoglycan-recognition protein lf;PDBTitle: crystal structure of the lfz ectodomain of the2 peptidoglycan recognition protein lf
9 d1ohta_
100.0
38
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like10 c1ohtA_
100.0
38
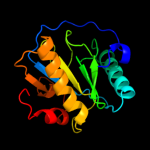
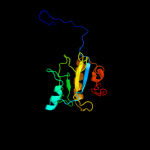
PDB header: hydrolaseChain: A: PDB Molecule: cg14704 protein;PDBTitle: peptidoglycan recognition protein-lb
11 d1sk4a_
100.0
77
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like12 c3ep1B_
100.0
31
PDB header: immune systemChain: B: PDB Molecule: pgrp-hd - peptidoglycan recognition proteinPDBTitle: structure of the pgrp-hd from alvinella pompejana
13 c2xz8A_
100.0
30
PDB header: immune systemChain: A: PDB Molecule: peptidoglycan-recognition protein lf;PDBTitle: crystal structure of the lfw ectodomain of the2 peptidoglycan recognition protein lf
14 d1lbaa_
100.0
27
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like15 d2bgxa2
99.9
15
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like16 d1yb0a1
99.9
11
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like17 c3hmaA_
99.9
22
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylmuramoyl-l-alanine amidase xlya;PDBTitle: amidase from bacillus subtilis
18 d1j3ga_
99.8
13
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like19 c4bolA_
99.8
21
PDB header: hydrolaseChain: A: PDB Molecule: ampdh2;PDBTitle: crystal structure of ampdh2 from pseudomonas aeruginosa in2 complex with pentapeptide
20 c2bh7A_
99.7
21
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylmuramoyl-l-alanine amidase;PDBTitle: crystal structure of a semet derivative of amid at 2.22 angstroms
21 c3latB_
not modelled
98.9
14
PDB header: hydrolaseChain: B: PDB Molecule: bifunctional autolysin;PDBTitle: crystal structure of staphylococcus peptidoglycan hydrolase2 amie
22 d2b3ya2
52.4
21
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain23 d1whqa_
not modelled
51.4
17
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)24 d2etla1
not modelled
48.3
24
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Ubiquitin carboxyl-terminal hydrolase UCH-L25 c2b3yB_
not modelled
45.6
21
PDB header: lyaseChain: B: PDB Molecule: iron-responsive element binding protein 1;PDBTitle: structure of a monoclinic crystal form of human cytosolic aconitase2 (irp1)
26 c4ig7A_
not modelled
33.0
19
PDB header: hydrolase/signaling proteinChain: A: PDB Molecule: ubiquitin c-terminal hydrolase 37;PDBTitle: crystal structure of trichinella spiralis uch37 bound to ubiquitin2 vinyl methyl ester
27 c2yzhD_
not modelled
25.6
24
PDB header: oxidoreductaseChain: D: PDB Molecule: probable thiol peroxidase;PDBTitle: crystal structure of peroxiredoxin-like protein from aquifex aeolicus
28 c2lrtA_
not modelled
25.6
24
PDB header: oxidoreductaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of the uncharacterized thioredoxin-like protein2 bvu_1432 from bacteroides vulgatus
29 d1xd3a_
not modelled
24.7
47
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Ubiquitin carboxyl-terminal hydrolase UCH-L30 d1a9xb1
not modelled
24.2
32
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Carbamoyl phosphate synthetase, small subunit N-terminal domainFamily: Carbamoyl phosphate synthetase, small subunit N-terminal domain31 c2ywiA_
not modelled
24.0
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical conserved protein;PDBTitle: crystal structure of uncharacterized conserved protein from2 geobacillus kaustophilus
32 d1cmxa_
not modelled
18.0
32
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Ubiquitin carboxyl-terminal hydrolase UCH-L33 d1zzoa1
not modelled
17.5
19
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like34 d1zj8a1
not modelled
17.1
22
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 335 c2k6xA_
not modelled
16.6
24
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma factor rpod;PDBTitle: autoregulation of a group 1 bacterial sigma factor involves2 the formation of a region 1.1- induced compacted structure
36 c3lorB_
not modelled
14.3
16
PDB header: isomeraseChain: B: PDB Molecule: thiol-disulfide isomerase and thioredoxins;PDBTitle: the crystal structure of a thiol-disulfide isomerase from2 corynebacterium glutamicum to 2.2a
37 c1keeH_
not modelled
13.5
32
PDB header: ligaseChain: H: PDB Molecule: carbamoyl-phosphate synthetase small chain;PDBTitle: inactivation of the amidotransferase activity of carbamoyl phosphate2 synthetase by the antibiotic acivicin
38 d1knga_
not modelled
13.1
19
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like39 d1t23a_
not modelled
13.1
43
Fold: Chromosomal protein MC1Superfamily: Chromosomal protein MC1Family: Chromosomal protein MC140 d1xm8a_
not modelled
12.9
13
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Glyoxalase II (hydroxyacylglutathione hydrolase)41 c3kebB_
not modelled
12.9
21
PDB header: oxidoreductaseChain: B: PDB Molecule: probable thiol peroxidase;PDBTitle: thiol peroxidase from chromobacterium violaceum
42 c1y88A_
not modelled
12.2
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein af1548;PDBTitle: crystal structure of protein of unknown function af1548
43 c3uinD_
not modelled
11.9
13
PDB header: ligase/isomerase/protein bindingChain: D: PDB Molecule: e3 sumo-protein ligase ranbp2;PDBTitle: complex between human rangap1-sumo2, ubc9 and the ir1 domain from2 ranbp2
44 c1u3eM_
not modelled
11.3
53
PDB header: dna binding protein/dnaChain: M: PDB Molecule: hnh homing endonuclease;PDBTitle: dna binding and cleavage by the hnh homing endonuclease i-hmui
45 c1r6tA_
not modelled
11.1
15
PDB header: ligaseChain: A: PDB Molecule: tryptophanyl-trna synthetase;PDBTitle: crystal structure of human tryptophanyl-trna synthetase
46 d1zofa1
not modelled
10.8
20
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like47 c2lvlA_
not modelled
10.5
36
PDB header: lantibiotic-binding-proteinChain: A: PDB Molecule: spai;PDBTitle: nmr structure the lantibiotic immunity protein spai
48 c3gknA_
not modelled
10.2
31
PDB header: oxidoreductaseChain: A: PDB Molecule: bacterioferritin comigratory protein;PDBTitle: insights into the alkyl peroxide reduction activity of xanthomonas2 campestris bacterioferritin comigratory protein from the trapped3 intermediate/ligand complex structures
49 c3t2dA_
not modelled
9.9
19
PDB header: lyase, hydrolaseChain: A: PDB Molecule: fructose-1,6-bisphosphate aldolase/phosphatase;PDBTitle: fructose-1,6-bisphosphate aldolase/phosphatase from thermoproteus2 neutrophilus, fbp-bound form
50 c2ywnA_
not modelled
9.9
23
PDB header: oxidoreductaseChain: A: PDB Molecule: peroxiredoxin-like protein;PDBTitle: crystal structure of peroxiredoxin-like protein from2 sulfolobus tokodaii
51 c2p5qA_
not modelled
9.6
47
PDB header: oxidoreductaseChain: A: PDB Molecule: glutathione peroxidase 5;PDBTitle: crystal structure of the poplar glutathione peroxidase 5 in2 the reduced form
52 c4fo5A_
not modelled
9.5
23
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin-like protein;PDBTitle: crystal structure of a thioredoxin-like protein (bdi_1100) from2 parabacteroides distasonis atcc 8503 at 2.02 a resolution
53 c4evmA_
not modelled
9.4
5
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin family protein;PDBTitle: 1.5 angstrom crystal structure of soluble domain of membrane-anchored2 thioredoxin family protein from streptococcus pneumoniae strain3 canada mdr_19a
54 c3cynC_
not modelled
9.2
25
PDB header: oxidoreductaseChain: C: PDB Molecule: probable glutathione peroxidase 8;PDBTitle: the structure of human gpx8
55 c3dwvB_
not modelled
9.2
25
PDB header: oxidoreductaseChain: B: PDB Molecule: glutathione peroxidase-like protein;PDBTitle: glutathione peroxidase-type tryparedoxin peroxidase, oxidized form
56 c2obiA_
not modelled
8.4
16
PDB header: oxidoreductaseChain: A: PDB Molecule: phospholipid hydroperoxide glutathionePDBTitle: crystal structure of the selenocysteine to cysteine mutant2 of human phospholipid hydroperoxide glutathione peroxidase3 (gpx4)
57 c3u5rG_
not modelled
8.4
21
PDB header: structural genomics, unknown functionChain: G: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a hypothetical protein smc02350 from2 sinorhizobium meliloti 1021
58 d1jfua_
not modelled
8.2
18
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like59 d2hq4a1
not modelled
8.1
14
Fold: PH1570-likeSuperfamily: PH1570-likeFamily: PH1570-like60 d1r6ta2
not modelled
8.1
15
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Class I aminoacyl-tRNA synthetases (RS), catalytic domain61 c1jrjA_
not modelled
8.0
16
PDB header: hormone/growth factorChain: A: PDB Molecule: exendin-4;PDBTitle: solution structure of exendin-4 in 30-vol% trifluoroethanol
62 c1d0rA_
not modelled
8.0
24
PDB header: hormone/growth factorChain: A: PDB Molecule: glucagon-like peptide-1-(7-36)-amide;PDBTitle: solution structure of glucagon-like peptide-1-(7-36)-amide2 in trifluoroethanol/water
63 d1gp1a_
not modelled
7.8
67
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like64 c3a7sA_
not modelled
7.7
24
PDB header: hydrolaseChain: A: PDB Molecule: ubiquitin carboxyl-terminal hydrolase isozyme l5;PDBTitle: catalytic domain of uch37
65 c2ls5A_
not modelled
7.5
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of a putative protein disulfide isomerase from2 bacteroides thetaiotaomicron
66 c2wdtA_
not modelled
7.4
21
PDB header: hydrolase/protein bindingChain: A: PDB Molecule: ubiquitin carboxyl-terminal hydrolase l3;PDBTitle: crystal structure of plasmodium falciparum uchl3 in complex2 with the suicide inhibitor ubvme
67 c1z5sD_
not modelled
7.2
13
PDB header: ligaseChain: D: PDB Molecule: ran-binding protein 2;PDBTitle: crystal structure of a complex between ubc9, sumo-1,2 rangap1 and nup358/ranbp2
68 d1xhoa_
not modelled
7.2
20
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase69 c1xhoB_
not modelled
7.2
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: chorismate mutase;PDBTitle: chorismate mutase from clostridium thermocellum cth-682
70 c2r37A_
not modelled
7.1
24
PDB header: oxidoreductaseChain: A: PDB Molecule: glutathione peroxidase 3;PDBTitle: crystal structure of human glutathione peroxidase 3 (selenocysteine to2 glycine mutant)
71 d2f8aa1
not modelled
6.8
67
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like72 c2l5oA_
not modelled
6.8
31
PDB header: oxidoreductaseChain: A: PDB Molecule: putative thioredoxin;PDBTitle: solution structure of a putative thioredoxin from neisseria2 meningitidis
73 c2kgsA_
not modelled
6.6
21
PDB header: membrane proteinChain: A: PDB Molecule: uncharacterized protein rv0899/mt0922;PDBTitle: solution structure of the amino-terminal domain of ompatb, a pore2 forming protein from mycobacterium tuberculosis
74 d1zyea1
not modelled
6.5
15
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like75 c4hdeA_
not modelled
6.5
38
PDB header: lipid binding proteinChain: A: PDB Molecule: sco1/senc family lipoprotein;PDBTitle: the crystal structure of a sco1/senc family lipoprotein from bacillus2 anthracis str. ames
76 c3tueB_
not modelled
6.5
20
PDB header: oxidoreductaseChain: B: PDB Molecule: tryparedoxin peroxidase;PDBTitle: the structure of tryparedoxin peroxidase i from leishmania major
77 c3ewlA_
not modelled
6.3
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized conserved protein bf1870;PDBTitle: crystal structure of conserved protein bf1870 of unknown function from2 bacteroides fragilis
78 d1vl6a1
not modelled
6.3
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain79 c2fyoA_
not modelled
6.3
21
PDB header: transferaseChain: A: PDB Molecule: carnitine o-palmitoyltransferase ii,PDBTitle: crystal structure of rat carnitine palmitoyltransferase 22 in space group p43212
80 c2ke4A_
not modelled
6.0
5
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
81 d1pj3a1
not modelled
6.0
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain82 c2b1kA_
not modelled
6.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein dsbe;PDBTitle: crystal structure of e. coli ccmg protein
83 c4eo3A_
not modelled
5.9
31
PDB header: oxidoreductaseChain: A: PDB Molecule: bacterioferritin comigratory protein/nadh dehydrogenase;PDBTitle: peroxiredoxin nitroreductase fusion enzyme
84 c2h66G_
not modelled
5.9
30
PDB header: structural genomics/oxidoreductaseChain: G: PDB Molecule: pv-pf14_0368;PDBTitle: the crystal structure of plasmodium vivax 2-cys2 peroxiredoxin
85 c2he3A_
not modelled
5.8
26
PDB header: oxidoreductaseChain: A: PDB Molecule: glutathione peroxidase 2;PDBTitle: crystal structure of the selenocysteine to cysteine mutant of human2 glutathionine peroxidase 2 (gpx2)
86 c3eurA_
not modelled
5.8
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the c-terminal domain of uncharacterized protein2 from bacteroides fragilis nctc 9343
87 c4ka0C_
not modelled
5.7
12
PDB header: oxidoreductaseChain: C: PDB Molecule: putative thiol-disulfide oxidoreductase;PDBTitle: crystal structure of a putative thiol-disulfide oxidoreductase from2 bacteroides vulgatus (target nysgrc-011676), space group p21221
88 d1m98a1
not modelled
5.6
14
Fold: Orange carotenoid protein, N-terminal domainSuperfamily: Orange carotenoid protein, N-terminal domainFamily: Orange carotenoid protein, N-terminal domain89 c4htyA_
not modelled
5.5
13
PDB header: hydrolaseChain: A: PDB Molecule: cellulase;PDBTitle: crystal structure of a metagenome-derived cellulase cel5a
90 d1o0sa1
not modelled
5.5
29
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain91 d1lu4a_
not modelled
5.5
10
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like92 c2p18A_
not modelled
5.5
11
PDB header: hydrolaseChain: A: PDB Molecule: glyoxalase ii;PDBTitle: crystal structure of the leishmania infantum glyoxalase ii
93 c2lrnA_
not modelled
5.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein;PDBTitle: solution structure of a thiol:disulfide interchange protein from2 bacteroides sp.
94 c2bmxB_
not modelled
5.3
14
PDB header: oxidoreductaseChain: B: PDB Molecule: alkyl hydroperoxidase c;PDBTitle: mycobacterium tuberculosis ahpc
95 c2p7jA_
not modelled
5.3
25
PDB header: transcriptionChain: A: PDB Molecule: putative sensory box/ggdef family protein;PDBTitle: crystal structure of the domain of putative sensory box/ggdef family2 protein from vibrio parahaemolyticus
96 c2m72A_
not modelled
5.3
31
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized thioredoxin-like protein;PDBTitle: solution structure of uncharacterized thioredoxin-like protein pg_21752 from porphyromonas gingivalis
97 c4je1A_
not modelled
5.3
11
PDB header: oxidoreductaseChain: A: PDB Molecule: probable thiol peroxidase;PDBTitle: crystal structure of thiol peroxidase from burkholderia cenocepacia2 j2315
98 d1gq2a1
not modelled
5.2
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain99 c2j8aA_
not modelled
5.2
18
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-4PDBTitle: x-ray structure of the n-terminus rrm domain of set1