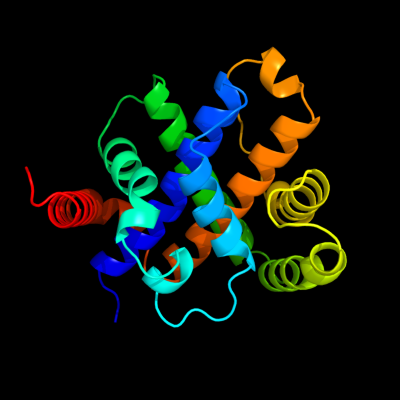
1 d1sdia_
100.0
100
Fold: YcfC-likeSuperfamily: YcfC-likeFamily: YcfC-like2 c2xr1A_
39.1
20
PDB header: hydrolaseChain: A: PDB Molecule: cleavage and polyadenylation specificity factor 100 kdPDBTitle: dimeric archaeal cleavage and polyadenylation specificity2 factor with n-terminal kh domains (kh-cpsf) from methanosarcina3 mazei
3 c2gejA_
33.1
12
PDB header: transferaseChain: A: PDB Molecule: phosphatidylinositol mannosyltransferase (pima);PDBTitle: crystal structure of phosphatidylinositol mannosyltransferase (pima)2 from mycobacterium smegmatis in complex with gdp-man
4 c2ycbA_
30.0
15
PDB header: hydrolaseChain: A: PDB Molecule: cleavage and polyadenylation specificity factor;PDBTitle: structure of the archaeal beta-casp protein with n-terminal2 kh domains from methanothermobacter thermautotrophicus
5 c2rdcA_
27.0
22
PDB header: lipid binding proteinChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative lipid binding protein (gsu0061) from2 geobacter sulfurreducens pca at 1.80 a resolution
6 c3af5A_
22.9
34
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein ph1404;PDBTitle: the crystal structure of an archaeal cpsf subunit, ph1404 from2 pyrococcus horikoshii
7 d2fgea1
21.5
17
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like8 c2k1vB_
15.7
46
PDB header: hormoneChain: B: PDB Molecule: relaxin-3;PDBTitle: r3/i5 relaxin chimera
9 c2rpaA_
15.1
28
PDB header: hydrolaseChain: A: PDB Molecule: katanin p60 atpase-containing subunit a1;PDBTitle: the solution structure of n-terminal domain of microtubule severing2 enzyme
10 c3fxdD_
14.0
36
PDB header: unknown functionChain: D: PDB Molecule: protein icmr;PDBTitle: crystal structure of interacting domains of icmr and icmq
11 c3cx3A_
11.7
7
PDB header: metal binding proteinChain: A: PDB Molecule: lipoprotein;PDBTitle: crystal structure analysis of the streptococcus pneumoniae2 adcaii protein
12 c3mzkC_
11.7
18
PDB header: protein transportChain: C: PDB Molecule: protein transport protein sec16;PDBTitle: sec13/sec16 complex, s.cerevisiae
13 c2rmsB_
11.6
56
PDB header: transcriptionChain: B: PDB Molecule: msin3a-binding protein;PDBTitle: solution structure of the msin3a pah1-sap25 sid complex
14 c2xdjF_
10.4
22
PDB header: unknown functionChain: F: PDB Molecule: uncharacterized protein ybgf;PDBTitle: crystal structure of the n-terminal domain of e.coli ybgf
15 c2h8bB_
9.8
40
PDB header: hormone/growth factorChain: B: PDB Molecule: insulin-like 3;PDBTitle: solution structure of insl3
16 c2ogwB_
9.0
13
PDB header: transport proteinChain: B: PDB Molecule: high-affinity zinc uptake system protein znuaPDBTitle: structure of abc type zinc transporter from e. coli
17 c2r5uD_
8.9
20
PDB header: hydrolaseChain: D: PDB Molecule: replicative dna helicase;PDBTitle: crystal structure of the n-terminal domain of dnab helicase from2 mycobacterium tuberculosis
18 c2jufA_
8.5
27
PDB header: gene regulationChain: A: PDB Molecule: p53-associated parkin-like cytoplasmic protein;PDBTitle: nmr solution structure of parc cph domain. nesg target2 hr3443b/sgc-toronto
19 d1qi9a_
8.3
29
Fold: Acid phosphatase/Vanadium-dependent haloperoxidaseSuperfamily: Acid phosphatase/Vanadium-dependent haloperoxidaseFamily: Haloperoxidase (bromoperoxidase)20 c2fhdA_
8.3
20
PDB header: cell cycleChain: A: PDB Molecule: dna repair protein rhp9/crb2;PDBTitle: crystal structure of crb2 tandem tudor domains
21 c3h25A_
not modelled
8.2
32
PDB header: replication/dnaChain: A: PDB Molecule: replication protein b;PDBTitle: crystal structure of the catalytic domain of primase repb' in complex2 with initiator dna
22 c3hgkE_
not modelled
8.2
20
PDB header: transferaseChain: E: PDB Molecule: effector protein hopab2;PDBTitle: crystal structure of effect protein avrptob complexed with2 kinase pto
23 d2jnga1
not modelled
7.9
27
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: CPH domain24 d1qhba_
not modelled
7.7
18
Fold: Acid phosphatase/Vanadium-dependent haloperoxidaseSuperfamily: Acid phosphatase/Vanadium-dependent haloperoxidaseFamily: Haloperoxidase (bromoperoxidase)25 d1vkea_
not modelled
7.6
13
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: CMD-like26 d1up8a_
not modelled
7.5
14
Fold: Acid phosphatase/Vanadium-dependent haloperoxidaseSuperfamily: Acid phosphatase/Vanadium-dependent haloperoxidaseFamily: Haloperoxidase (bromoperoxidase)27 c2o1eB_
not modelled
7.5
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ycdh;PDBTitle: crystal structure of the metal-dependent lipoprotein ycdh2 from bacillus subtilis, northeast structural genomics3 target sr583
28 d1u7ka_
not modelled
7.2
33
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain29 d2jeka1
not modelled
7.1
20
Fold: Rv1873-likeSuperfamily: Rv1873-likeFamily: Rv1873-like30 c1yv0T_
not modelled
7.0
27
PDB header: contractile proteinChain: T: PDB Molecule: troponin t, fast skeletal muscle isoforms;PDBTitle: crystal structure of skeletal muscle troponin in the ca2+-2 free state
31 c1xouA_
not modelled
7.0
24
PDB header: structural protein/chaperoneChain: A: PDB Molecule: espa;PDBTitle: crystal structure of the cesa-espa complex
32 d1xoua_
not modelled
7.0
24
Fold: EspA/CesA-likeSuperfamily: EspA/CesA-likeFamily: EspA-like33 d2v7qi1
not modelled
6.7
60
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Epsilon subunit of mitochondrial F1F0-ATP synthaseFamily: Epsilon subunit of mitochondrial F1F0-ATP synthase34 c2w9zA_
not modelled
6.5
17
PDB header: transferaseChain: A: PDB Molecule: g1/s-specific cyclin-d1;PDBTitle: crystal structure of cdk4 in complex with a d-type cyclin
35 c3nrhA_
not modelled
6.4
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein bf1032;PDBTitle: crystal structure of protein bf1032 from bacteroides fragilis,2 northeast structural genomics consortium target bfr309
36 d1jwea_
not modelled
6.2
18
Fold: N-terminal domain of DnaB helicaseSuperfamily: N-terminal domain of DnaB helicaseFamily: N-terminal domain of DnaB helicase37 c1cosA_
not modelled
6.2
29
PDB header: alpha-helical bundleChain: A: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-2 helical bundle
38 c1cosB_
not modelled
6.2
29
PDB header: alpha-helical bundleChain: B: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-2 helical bundle
39 c1cosC_
not modelled
6.2
29
PDB header: alpha-helical bundleChain: C: PDB Molecule: coiled serine;PDBTitle: crystal structure of a synthetic triple-stranded alpha-2 helical bundle
40 c3imoC_
not modelled
6.1
21
PDB header: unknown functionChain: C: PDB Molecule: integron cassette protein;PDBTitle: structure from the mobile metagenome of vibrio cholerae.2 integron cassette protein vch_cass14
41 d1hula_
not modelled
5.7
19
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Short-chain cytokines42 c2f59B_
not modelled
5.5
20
PDB header: transferaseChain: B: PDB Molecule: 6,7-dimethyl-8-ribityllumazine synthase 1;PDBTitle: lumazine synthase ribh1 from brucella abortus (gene bruab1_0785,2 swiss-prot entry q57dy1) complexed with inhibitor 5-nitro-6-(d-3 ribitylamino)-2,4(1h,3h) pyrimidinedione
43 d1b8za_
not modelled
5.5
14
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein44 d2bs2b2
not modelled
5.4
24
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins45 c3dlaD_
not modelled
5.3
19
PDB header: ligaseChain: D: PDB Molecule: glutamine-dependent nad(+) synthetase;PDBTitle: x-ray crystal structure of glutamine-dependent nad+ synthetase from2 mycobacterium tuberculosis bound to naad+ and don
46 c3c4vB_
not modelled
5.2
14
PDB header: transferaseChain: B: PDB Molecule: predicted glycosyltransferases;PDBTitle: structure of the retaining glycosyltransferase msha:the2 first step in mycothiol biosynthesis. organism:3 corynebacterium glutamicum : complex with udp and 1l-ins-1-4 p.
47 c3lk2B_
not modelled
5.0
18
PDB header: protein bindingChain: B: PDB Molecule: f-actin-capping protein subunit beta isoforms 1 and 2;PDBTitle: crystal structure of capz bound to the uncapping motif from carmil