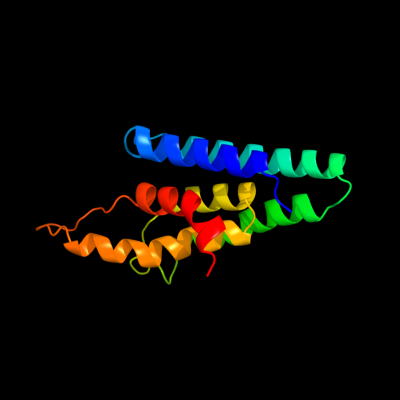
1 c3s0xB_
98.7
19
PDB header: hydrolaseChain: B: PDB Molecule: peptidase a24b, flak domain protein;PDBTitle: the crystal structure of gxgd membrane protease flak
2 c2owoA_
86.6
16
PDB header: ligase/dnaChain: A: PDB Molecule: dna ligase;PDBTitle: last stop on the road to repair: structure of e.coli dna ligase bound2 to nicked dna-adenylate
3 c1v9pB_
81.9
12
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase
4 c2k5cA_
81.7
25
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein pf0385;PDBTitle: nmr structure for pf0385
5 c3py7A_
78.6
21
PDB header: viral proteinChain: A: PDB Molecule: maltose-binding periplasmic protein,paxillin ld1,protein e6PDBTitle: crystal structure of full-length bovine papillomavirus oncoprotein e62 in complex with ld1 motif of paxillin at 2.3a resolution
6 c1dgsB_
76.6
17
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase from t.2 filiformis
7 d1dgsa1
74.9
27
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: NAD+-dependent DNA ligase, domain 38 d2fk4a1
69.5
19
Fold: E6 C-terminal domain-likeSuperfamily: E6 C-terminal domain-likeFamily: E6 C-terminal domain-like9 d1vd4a_
69.2
27
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain10 d2fiya1
66.5
22
Fold: FdhE-likeSuperfamily: FdhE-likeFamily: FdhE-like11 c2qgpA_
66.5
18
PDB header: hydrolaseChain: A: PDB Molecule: hnh endonuclease;PDBTitle: x-ray structure of the nhn endonuclease from geobacter2 metallireducens. northeast structural genomics consortium3 target gmr87.
12 c3zyqA_
64.9
18
PDB header: signalingChain: A: PDB Molecule: hepatocyte growth factor-regulated tyrosine kinasePDBTitle: crystal structure of the tandem vhs and fyve domains of hepatocyte2 growth factor-regulated tyrosine kinase substrate (hgs-hrs) at 1.483 a resolution
13 d1vfya_
63.6
29
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: FYVE, a phosphatidylinositol-3-phosphate binding domain14 d1pfva3
63.0
27
Fold: Rubredoxin-likeSuperfamily: Methionyl-tRNA synthetase (MetRS), Zn-domainFamily: Methionyl-tRNA synthetase (MetRS), Zn-domain15 c2ea6A_
62.8
21
PDB header: transcriptionChain: A: PDB Molecule: ring finger protein 4;PDBTitle: solution structure of the ring domain of the human ring2 finger protein 4
16 d2jnea1
62.1
43
Fold: Rubredoxin-likeSuperfamily: YfgJ-likeFamily: YfgJ-like17 c2jneA_
62.1
43
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein yfgj;PDBTitle: nmr structure of e.coli yfgj modelled with two zn+2 bound.2 northeast structural genomics consortium target er317.
18 c2lcqA_
62.0
31
PDB header: metal binding proteinChain: A: PDB Molecule: putative toxin vapc6;PDBTitle: solution structure of the endonuclease nob1 from p.horikoshii
19 d1wfpa_
61.6
18
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger20 d1dvpa2
59.7
19
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: FYVE, a phosphatidylinositol-3-phosphate binding domain21 c3nw0A_
not modelled
59.4
18
PDB header: metal binding proteinChain: A: PDB Molecule: non-structural maintenance of chromosomes element 1PDBTitle: crystal structure of mageg1 and nse1 complex
22 c3llkA_
not modelled
58.7
23
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfhydryl oxidase 1;PDBTitle: sulfhydryl oxidase fragment of human qsox1
23 d1wg2a_
not modelled
58.6
18
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger24 c1dvpA_
not modelled
58.5
24
PDB header: transferaseChain: A: PDB Molecule: hepatocyte growth factor-regulated tyrosinePDBTitle: crystal structure of the vhs and fyve tandem domains of hrs,2 a protein involved in membrane trafficking and signal3 transduction
25 d1wfka_
not modelled
57.6
36
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: FYVE, a phosphatidylinositol-3-phosphate binding domain26 c2kr6A_
not modelled
57.4
21
PDB header: hydrolaseChain: A: PDB Molecule: presenilin-1;PDBTitle: solution structure of presenilin-1 ctf subunit
27 c3floD_
55.8
18
PDB header: transferaseChain: D: PDB Molecule: dna polymerase alpha catalytic subunit a;PDBTitle: crystal structure of the carboxyl-terminal domain of yeast2 dna polymerase alpha in complex with its b subunit
28 c3pfqA_
not modelled
55.7
18
PDB header: transferaseChain: A: PDB Molecule: protein kinase c beta type;PDBTitle: crystal structure and allosteric activation of protein kinase c beta2 ii
29 c2ecmA_
not modelled
55.1
26
PDB header: metal binding proteinChain: A: PDB Molecule: ring finger and chy zinc finger domain-PDBTitle: solution structure of the ring domain of the ring finger2 and chy zinc finger domain-containing protein 1 from mus3 musculus
30 c3odeA_
not modelled
54.3
33
PDB header: dna binding protein/dnaChain: A: PDB Molecule: poly [adp-ribose] polymerase 1;PDBTitle: human parp-1 zinc finger 2 (zn2) bound to dna
31 c3k1lA_
not modelled
54.3
43
PDB header: ligaseChain: A: PDB Molecule: fancl;PDBTitle: crystal structure of fancl
32 c2dmjA_
not modelled
54.2
25
PDB header: transferaseChain: A: PDB Molecule: poly (adp-ribose) polymerase family, member 1;PDBTitle: solution structure of the first zf-parp domain of human2 poly(adp-ribose)polymerase-1
33 c2hr5B_
not modelled
53.6
17
PDB header: metal binding proteinChain: B: PDB Molecule: rubrerythrin;PDBTitle: pf1283- rubrerythrin from pyrococcus furiosus iron bound form
34 c2ep4A_
not modelled
53.6
21
PDB header: protein bindingChain: A: PDB Molecule: ring finger protein 24;PDBTitle: solution structure of ring finger from human ring finger2 protein 24
35 d1wfha_
not modelled
53.5
26
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger36 c2kizA_
not modelled
52.8
26
PDB header: metal binding proteinChain: A: PDB Molecule: e3 ubiquitin-protein ligase arkadia;PDBTitle: solution structure of arkadia ring-h2 finger domain
37 c2jrpA_
not modelled
52.6
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative cytoplasmic protein;PDBTitle: solution nmr structure of yfgj from salmonella typhimurium2 modeled with two zn+2 bound, northeast structural genomics3 consortium target str86
38 d2baya1
not modelled
50.4
11
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: U-box39 d2gmga1
not modelled
50.3
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PF0610-like40 c1x4wA_
not modelled
50.2
35
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein flj13222;PDBTitle: solution structure of the zf-an1 domain from human2 hypothetical protein flj13222
41 c1z2qA_
not modelled
49.8
16
PDB header: membrane proteinChain: A: PDB Molecule: lm5-1;PDBTitle: high-resolution solution structure of the lm5-1 fyve domain2 from leishmania major
42 c2l0bA_
not modelled
49.6
25
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase praja-1;PDBTitle: solution nmr structure of zinc finger domain of e3 ubiquitin-protein2 ligase praja-1 from homo sapiens, northeast structural genomics3 consortium (nesg) target hr4710b
43 c2ja6L_
not modelled
49.4
28
PDB header: transferaseChain: L: PDB Molecule: dna-directed rna polymerases i, ii, and iii 7.7PDBTitle: cpd lesion containing rna polymerase ii elongation complex2 b
44 c2k9hA_
not modelled
48.4
17
PDB header: metal binding proteinChain: A: PDB Molecule: glycoprotein;PDBTitle: the hantavirus glycoprotein g1 tail contains a dual cchc-2 type classical zinc fingers
45 c2d8tA_
not modelled
47.4
22
PDB header: metal binding proteinChain: A: PDB Molecule: ring finger protein 146;PDBTitle: solution structure of the ring domain of the human ring2 finger protein 146
46 d1iyma_
not modelled
46.9
20
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: RING finger domain, C3HC447 c2ct0A_
not modelled
46.6
20
PDB header: gene regulationChain: A: PDB Molecule: non-smc element 1 homolog;PDBTitle: solution structure of the ring domain of the non-smc2 element 1 protein
48 c2kdxA_
not modelled
46.5
25
PDB header: metal-binding proteinChain: A: PDB Molecule: hydrogenase/urease nickel incorporation proteinPDBTitle: solution structure of hypa protein
49 c2vjfB_
not modelled
46.4
16
PDB header: ligaseChain: B: PDB Molecule: mdm4 protein;PDBTitle: crystal structure of the mdm2-mdmx ring domain heterodimer
50 c1dvbA_
not modelled
46.0
17
PDB header: electron transportChain: A: PDB Molecule: rubrerythrin;PDBTitle: rubrerythrin
51 c3axtA_
not modelled
45.4
30
PDB header: transferaseChain: A: PDB Molecule: probable n(2),n(2)-dimethylguanosine trna methyltransferasePDBTitle: complex structure of trna methyltransferase trm1 from aquifex aeolicus2 with s-adenosyl-l-methionine
52 c2cs2A_
not modelled
45.2
33
PDB header: transferaseChain: A: PDB Molecule: poly [adp-ribose] polymerase-1;PDBTitle: solution structure of the second zn-finger domain of2 poly(adp-ribose) polymerase-1
53 c3a44D_
not modelled
44.6
14
PDB header: metal binding proteinChain: D: PDB Molecule: hydrogenase nickel incorporation protein hypa;PDBTitle: crystal structure of hypa in the dimeric form
54 c3h0gL_
not modelled
44.4
28
PDB header: transcriptionChain: L: PDB Molecule: dna-directed rna polymerases i, ii, and iiiPDBTitle: rna polymerase ii from schizosaccharomyces pombe
55 c3bjiA_
not modelled
44.0
31
PDB header: signaling proteinChain: A: PDB Molecule: proto-oncogene vav;PDBTitle: structural basis of promiscuous guanine nucleotide exchange2 by the t-cell essential vav1
56 c3ky9B_
not modelled
43.9
29
PDB header: apoptosisChain: B: PDB Molecule: proto-oncogene vav;PDBTitle: autoinhibited vav1
57 d2ctda2
not modelled
43.9
44
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H258 d2gnra1
not modelled
43.8
29
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: SSO2064-like59 c2xeuA_
not modelled
43.7
26
PDB header: transcriptionChain: A: PDB Molecule: ring finger protein 4;PDBTitle: ring domain
60 d1rmda2
not modelled
43.7
35
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: RING finger domain, C3HC461 d2ayja1
not modelled
43.6
17
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L40e62 c1v9xA_
not modelled
42.7
25
PDB header: transferaseChain: A: PDB Molecule: poly (adp-ribose) polymerase;PDBTitle: solution structure of the first zn-finger domain of2 poly(adp-ribose) polymerase-1
63 c2djbA_
not modelled
42.5
16
PDB header: gene regulationChain: A: PDB Molecule: polycomb group ring finger protein 6;PDBTitle: solution structure of the ring domain of the human polycomb2 group ring finger protein 6
64 c1x4jA_
not modelled
42.0
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ring finger protein 38;PDBTitle: solution structure of ring finger in ring finger protein 38
65 c2qkdA_
not modelled
41.9
24
PDB header: signaling protein, cell cycleChain: A: PDB Molecule: zinc finger protein zpr1;PDBTitle: crystal structure of tandem zpr1 domains
66 c2f9iD_
not modelled
41.9
16
PDB header: transferaseChain: D: PDB Molecule: acetyl-coenzyme a carboxylase carboxylPDBTitle: crystal structure of the carboxyltransferase subunit of acc2 from staphylococcus aureus
67 d1faqa_
not modelled
41.4
37
Fold: Cysteine-rich domainSuperfamily: Cysteine-rich domainFamily: Protein kinase cysteine-rich domain (cys2, phorbol-binding domain)68 c2ectA_
not modelled
41.4
19
PDB header: metal binding proteinChain: A: PDB Molecule: ring finger protein 126;PDBTitle: solution structure of the zinc finger, c3hc4 type (ring2 finger) domain of ring finger protein 126
69 c2i5oA_
not modelled
41.2
25
PDB header: transferaseChain: A: PDB Molecule: dna polymerase eta;PDBTitle: solution structure of the ubiquitin-binding zinc finger2 (ubz) domain of the human dna y-polymerase eta
70 c3cngC_
not modelled
41.0
20
PDB header: hydrolaseChain: C: PDB Molecule: nudix hydrolase;PDBTitle: crystal structure of nudix hydrolase from nitrosomonas europaea
71 c2f42A_
not modelled
40.8
17
PDB header: chaperoneChain: A: PDB Molecule: stip1 homology and u-box containing protein 1;PDBTitle: dimerization and u-box domains of zebrafish c-terminal of hsp702 interacting protein
72 c3lrqB_
not modelled
40.7
32
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase trim37;PDBTitle: crystal structure of the u-box domain of human ubiquitin-2 protein ligase (e3), northeast structural genomics3 consortium target hr4604d.
73 d1uw0a_
not modelled
40.6
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: PARP-type zinc finger74 c2zkr2_
not modelled
40.4
23
PDB header: ribosomal protein/rnaChain: 2: PDB Molecule: 60s ribosomal protein l37e;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
75 d1vqo11
not modelled
39.5
30
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37e76 d1wjva1
not modelled
39.2
26
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: C2HC finger77 c2rowA_
not modelled
39.1
35
PDB header: transferaseChain: A: PDB Molecule: rho-associated protein kinase 2;PDBTitle: the c1 domain of rock ii
78 c3tsuA_
not modelled
38.8
12
PDB header: transferaseChain: A: PDB Molecule: transcriptional regulatory protein;PDBTitle: crystal structure of e. coli hypf with amp-pnp and carbamoyl phosphate
79 d1wffa_
not modelled
38.5
28
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger80 c3t7lA_
not modelled
38.5
22
PDB header: transport proteinChain: A: PDB Molecule: zinc finger fyve domain-containing protein 16;PDBTitle: crystal structure of the fyve domain of endofin (zfyve16) at 1.1a2 resolution
81 d2cu8a1
not modelled
38.4
36
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain82 d2ct1a1
not modelled
38.1
60
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H283 c2jr6A_
not modelled
37.7
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0434 protein nma0874;PDBTitle: solution structure of upf0434 protein nma0874. northeast structural2 genomics target mr32
84 c2pmzB_
not modelled
37.6
19
PDB header: translation, transferaseChain: B: PDB Molecule: dna-directed rna polymerase subunit b;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
85 c2jmdA_
not modelled
37.4
17
PDB header: ligaseChain: A: PDB Molecule: tnf receptor-associated factor 6;PDBTitle: solution structure of the ring domain of human traf6
86 c3jywY_
not modelled
35.8
36
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l37(a);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
87 d1tjla2
not modelled
35.7
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Prokaryotic DksA/TraR C4-type zinc finger88 d1q1aa_
not modelled
35.5
17
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators89 d1xa6a3
not modelled
35.4
35
Fold: Cysteine-rich domainSuperfamily: Cysteine-rich domainFamily: Protein kinase cysteine-rich domain (cys2, phorbol-binding domain)90 c1jocA_
not modelled
35.3
28
PDB header: membrane proteinChain: A: PDB Molecule: early endosomal autoantigen 1;PDBTitle: eea1 homodimer of c-terminal fyve domain bound to inositol2 1,3-diphosphate
91 c2y43B_
not modelled
35.0
14
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase rad18;PDBTitle: rad18 ubiquitin ligase ring domain structure
92 c1hk8A_
not modelled
33.7
27
PDB header: oxidoreductaseChain: A: PDB Molecule: anaerobic ribonucleotide-triphosphate reductase;PDBTitle: structural basis for allosteric substrate specificity2 regulation in class iii ribonucleotide reductases:3 nrdd in complex with dgtp
93 d1hk8a_
not modelled
33.7
27
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: Class III anaerobic ribonucleotide reductase NRDD subunit94 c3t6pA_
not modelled
33.7
40
PDB header: apoptosisChain: A: PDB Molecule: baculoviral iap repeat-containing protein 2;PDBTitle: iap antagonist-induced conformational change in ciap1 promotes e32 ligase activation via dimerization
95 c1s1iY_
not modelled
33.6
36
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l37-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
96 c2db6A_
not modelled
33.5
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sh3 and cysteine rich domain 3;PDBTitle: solution structure of rsgi ruh-051, a c1 domain of stac32 from human cdna
97 d1s5pa_
not modelled
32.9
8
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators98 c2csyA_
not modelled
32.8
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: zinc finger protein 183-like 1;PDBTitle: solution structure of the ring domain of the zinc finger2 protein 183-like 1
99 d2cona1
not modelled
32.7
36
Fold: Rubredoxin-likeSuperfamily: NOB1 zinc finger-likeFamily: NOB1 zinc finger-like100 d1chca_
not modelled
32.3
16
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: RING finger domain, C3HC4101 c2eluA_
not modelled
32.0
50
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 5th c2h2 zinc finger of human2 zinc finger protein 406
102 c2ecvA_
not modelled
31.5
24
PDB header: ligaseChain: A: PDB Molecule: tripartite motif-containing protein 5;PDBTitle: solution structure of the zinc finger, c3hc4 type (ring2 finger) domain of tripartite motif-containing protein 5
103 d1zbdb_
not modelled
31.4
18
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: FYVE, a phosphatidylinositol-3-phosphate binding domain104 c2eciA_
not modelled
31.4
16
PDB header: metal binding proteinChain: A: PDB Molecule: tnf receptor-associated factor 6;PDBTitle: solution structure of the ring domain of the human tnf2 receptor-associated factor 6 protein
105 c1u5kA_
not modelled
31.0
18
PDB header: recombination,replicationChain: A: PDB Molecule: hypothetical protein;PDBTitle: recombinational repair protein reco
106 c2kgoA_
not modelled
30.9
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ybii;PDBTitle: solution nmr structure of zn finger protein ybil from escherichia2 coli. nesg target et107, ocsp target ec0402
107 c1z6uA_
not modelled
30.8
19
PDB header: ligaseChain: A: PDB Molecule: np95-like ring finger protein isoform b;PDBTitle: np95-like ring finger protein isoform b [homo sapiens]
108 d1jm7a_
not modelled
30.8
32
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: RING finger domain, C3HC4109 d2c2la2
not modelled
30.7
17
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: U-box110 c1tjlD_
not modelled
30.7
33
PDB header: transcriptionChain: D: PDB Molecule: dnak suppressor protein;PDBTitle: crystal structure of transcription factor dksa from e. coli
111 d1wfla_
not modelled
29.9
19
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger112 c3u50C_
not modelled
29.9
19
PDB header: dna binding proteinChain: C: PDB Molecule: telomerase-associated protein 82;PDBTitle: crystal structure of the tetrahymena telomerase processivity factor2 teb1 ob-c
113 d1x3ha1
not modelled
29.7
23
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain114 c2hcnA_
not modelled
29.5
22
PDB header: transferaseChain: A: PDB Molecule: rna-directed rna polymerase (ns5);PDBTitle: crystal structure of rna dependent rna polymerase domain from west2 nile virus
115 d1iyca_
not modelled
29.4
35
Fold: Invertebrate chitin-binding proteinsSuperfamily: Invertebrate chitin-binding proteinsFamily: Antifungal peptide scarabaecin116 c2cszA_
not modelled
29.0
19
PDB header: signaling proteinChain: A: PDB Molecule: synaptotagmin-like protein 4;PDBTitle: solution structure of the ring domain of the synaptotagmin-2 like protein 4
117 d1kbea_
not modelled
28.9
33
Fold: Cysteine-rich domainSuperfamily: Cysteine-rich domainFamily: Protein kinase cysteine-rich domain (cys2, phorbol-binding domain)118 d1yc5a1
not modelled
28.7
23
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators119 c2klxA_
not modelled
28.5
23
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaredoxin;PDBTitle: solution structure of glutaredoxin from bartonella henselae str.2 houston
120 d1dsva_
not modelled
28.4
29
Fold: Retrovirus zinc finger-like domainsSuperfamily: Retrovirus zinc finger-like domainsFamily: Retrovirus zinc finger-like domains