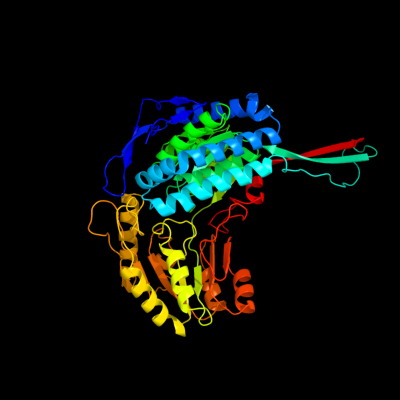
1 c2o2qA_
100.0
37
PDB header: oxidoreductaseChain: A: PDB Molecule: formyltetrahydrofolate dehydrogenase;PDBTitle: crystal structure of the c-terminal domain of rat2 10'formyltetrahydrofolate dehydrogenase in complex with nadp
2 d1bxsa_
100.0
37
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like3 c3ed6B_
100.0
37
PDB header: oxidoreductaseChain: B: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: 1.7 angstrom resolution crystal structure of betaine aldehyde2 dehydrogenase (betb) from staphylococcus aureus
4 c2jg7G_
100.0
28
PDB header: oxidoreductaseChain: G: PDB Molecule: antiquitin;PDBTitle: crystal structure of seabream antiquitin and elucidation of2 its substrate specificity
5 d1o9ja_
100.0
38
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like6 d1o04a_
100.0
40
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like7 c3qanB_
100.0
31
PDB header: oxidoreductaseChain: B: PDB Molecule: 1-pyrroline-5-carboxylate dehydrogenase 1;PDBTitle: crystal structure of 1-pyrroline-5-carboxylate dehydrogenase from2 bacillus halodurans
8 c2d4eB_
100.0
36
PDB header: oxidoreductaseChain: B: PDB Molecule: 5-carboxymethyl-2-hydroxymuconate semialdehydePDBTitle: crystal structure of the hpcc from thermus thermophilus hb8
9 c3iwkB_
100.0
36
PDB header: oxidoreductaseChain: B: PDB Molecule: aminoaldehyde dehydrogenase;PDBTitle: crystal structure of aminoaldehyde dehydrogenase 1 from2 pisum sativum (psamadh1)
10 d1a4sa_
100.0
35
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like11 d1ag8a_
100.0
40
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like12 d1uzba_
100.0
31
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like13 c2ve5H_
100.0
41
PDB header: oxidoreductaseChain: H: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: crystallographic structure of betaine aldehyde2 dehydrogenase from pseudomonas aeruginosa
14 c3rh9A_
100.0
32
PDB header: oxidoreductaseChain: A: PDB Molecule: succinate-semialdehyde dehydrogenase (nad(p)(+));PDBTitle: the crystal structure of oxidoreductase from marinobacter aquaeolei
15 d1wnda_
100.0
38
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like16 c3b4wA_
100.0
35
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of mycobacterium tuberculosis aldehyde dehydrogenase2 complexed with nad+
17 c3ifgH_
100.0
37
PDB header: oxidoreductaseChain: H: PDB Molecule: succinate-semialdehyde dehydrogenase (nadp+);PDBTitle: crystal structure of succinate-semialdehyde dehydrogenase from2 burkholderia pseudomallei, part 1 of 2
18 c3ek1C_
100.0
34
PDB header: oxidoreductaseChain: C: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from brucella2 melitensis biovar abortus 2308
19 d1euha_
100.0
33
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like20 c3r31A_
100.0
38
PDB header: oxidoreductaseChain: A: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: crystal structure of betaine aldehyde dehydrogenase from agrobacterium2 tumefaciens
21 c3k2wD_
not modelled
100.0
34
PDB header: oxidoreductaseChain: D: PDB Molecule: betaine-aldehyde dehydrogenase;PDBTitle: crystal structure of betaine-aldehyde dehydrogenase from2 pseudoalteromonas atlantica t6c
22 c3jz4C_
not modelled
100.0
35
PDB header: oxidoreductaseChain: C: PDB Molecule: succinate-semialdehyde dehydrogenase [nadp+];PDBTitle: crystal structure of e. coli nadp dependent enzyme
23 c1t90B_
not modelled
100.0
30
PDB header: oxidoreductaseChain: B: PDB Molecule: probable methylmalonate-semialdehydePDBTitle: crystal structure of methylmalonate semialdehyde2 dehydrogenase from bacillus subtilis
24 c2w8qA_
not modelled
100.0
34
PDB header: oxidoreductaseChain: A: PDB Molecule: succinate-semialdehyde dehydrogenase,PDBTitle: the crystal structure of human ssadh in complex with ssa.
25 c2hg2A_
not modelled
100.0
33
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase a;PDBTitle: structure of lactaldehyde dehydrogenase
26 c3i44A_
not modelled
100.0
34
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from bartonella2 henselae at 2.0a resolution
27 d1ky8a_
not modelled
100.0
27
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like28 c3prlD_
not modelled
100.0
33
PDB header: oxidoreductaseChain: D: PDB Molecule: nadp-dependent glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of nadp-dependent glyceraldehyde-3-phosphate2 dehydrogenase from bacillus halodurans c-125
29 d1bi9a_
not modelled
100.0
37
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like30 c3hazA_
not modelled
100.0
30
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase;PDBTitle: crystal structure of bifunctional proline utilization a2 (puta) protein
31 c3ju8B_
not modelled
100.0
28
PDB header: oxidoreductaseChain: B: PDB Molecule: succinylglutamic semialdehyde dehydrogenase;PDBTitle: crystal structure of succinylglutamic semialdehyde dehydrogenase from2 pseudomonas aeruginosa.
32 c3rosA_
not modelled
100.0
28
PDB header: oxidoreductaseChain: A: PDB Molecule: nad-dependent aldehyde dehydrogenase;PDBTitle: crystal structure of nad-dependent aldehyde dehydrogenase from2 lactobacillus acidophilus
33 c3efvC_
not modelled
100.0
31
PDB header: oxidoreductaseChain: C: PDB Molecule: putative succinate-semialdehyde dehydrogenase;PDBTitle: crystal structure of a putative succinate-semialdehyde dehydrogenase2 from salmonella typhimurium lt2 with bound nad
34 c2vroB_
not modelled
100.0
21
PDB header: oxidoreductaseChain: B: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from2 burkholderia xenovorans lb400
35 c3r64A_
not modelled
100.0
30
PDB header: oxidoreductaseChain: A: PDB Molecule: nad dependent benzaldehyde dehydrogenase;PDBTitle: crystal structure of a nad-dependent benzaldehyde dehydrogenase from2 corynebacterium glutamicum
36 c3pqaA_
not modelled
100.0
31
PDB header: oxidoreductaseChain: A: PDB Molecule: lactaldehyde dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase gapn2 from methanocaldococcus jannaschii dsm 2661
37 d1ad3a_
not modelled
100.0
27
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like38 c3v4cB_
not modelled
100.0
26
PDB header: oxidoreductaseChain: B: PDB Molecule: aldehyde dehydrogenase (nadp+);PDBTitle: crystal structure of a semialdehyde dehydrogenase from sinorhizobium2 meliloti 1021
39 d1ez0a_
not modelled
100.0
22
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like40 c3lnsD_
not modelled
100.0
28
PDB header: oxidoreductaseChain: D: PDB Molecule: benzaldehyde dehydrogenase;PDBTitle: benzaldehyde dehydrogenase, a class 3 aldehyde dehydrogenase, with2 bound nadp+ and benzoate adduct
41 c3k9dD_
not modelled
100.0
20
PDB header: oxidoreductaseChain: D: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of probable aldehyde dehydrogenase from listeria2 monocytogenes egd-e
42 d1o20a_
not modelled
100.0
14
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like43 c3my7A_
not modelled
100.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: alcohol dehydrogenase/acetaldehyde dehydrogenase;PDBTitle: the crystal structure of the acdh domain of an alcohol dehydrogenase2 from vibrio parahaemolyticus to 2.25a
44 c2h5gA_
not modelled
100.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: delta 1-pyrroline-5-carboxylate synthetase;PDBTitle: crystal structure of human pyrroline-5-carboxylate synthetase
45 d1vlua_
not modelled
100.0
16
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like46 c1vluB_
not modelled
100.0
15
PDB header: oxidoreductaseChain: B: PDB Molecule: gamma-glutamyl phosphate reductase;PDBTitle: crystal structure of gamma-glutamyl phosphate reductase (yor323c) from2 saccharomyces cerevisiae at 2.40 a resolution
47 d1k75a_
not modelled
98.8
18
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: L-histidinol dehydrogenase HisD48 c2yvqA_
not modelled
52.2
13
PDB header: ligaseChain: A: PDB Molecule: carbamoyl-phosphate synthase;PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
49 d1s7ia_
not modelled
50.6
20
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: DGPF domain (Pfam 04946)50 d1a9xa2
not modelled
43.7
22
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain51 d1wo8a1
not modelled
33.5
16
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Methylglyoxal synthase, MgsA52 c3jtpB_
not modelled
32.6
17
PDB header: protein bindingChain: B: PDB Molecule: adapter protein meca 1;PDBTitle: crystal structure of the c-terminal domain of meca
53 d1y5ea1
not modelled
28.9
23
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like54 c3e5bB_
not modelled
24.6
14
PDB header: lyaseChain: B: PDB Molecule: isocitrate lyase;PDBTitle: 2.4 a crystal structure of isocitrate lyase from brucella2 melitensis
55 d1o66a_
not modelled
24.2
10
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Ketopantoate hydroxymethyltransferase PanB56 c2yukA_
not modelled
19.7
19
PDB header: transferaseChain: A: PDB Molecule: myeloid/lymphoid or mixed-lineage leukemiaPDBTitle: solution structure of the hmg box of human myeloid/lymphoid2 or mixed-lineage leukemia protein 3 homolog
57 d2ioja1
not modelled
19.2
17
Fold: MurF and HprK N-domain-likeSuperfamily: HprK N-terminal domain-likeFamily: DRTGG domain58 d1mkza_
not modelled
16.9
20
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like59 d1u0ta_
not modelled
15.3
20
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: NAD kinase-like60 d1js8a1
not modelled
14.7
11
Fold: Di-copper centre-containing domainSuperfamily: Di-copper centre-containing domainFamily: Hemocyanin middle domain61 d2bona1
not modelled
14.1
16
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: Diacylglycerol kinase-like62 d1h6da1
not modelled
13.0
6
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain63 c3v4gA_
not modelled
12.5
24
PDB header: dna binding proteinChain: A: PDB Molecule: arginine repressor;PDBTitle: 1.60 angstrom resolution crystal structure of an arginine repressor2 from vibrio vulnificus cmcp6
64 c2ec4A_
not modelled
12.1
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: fas-associated factor 1;PDBTitle: solution structure of the uas domain from human fas-2 associated factor 1
65 d1xxaa_
not modelled
12.1
24
Fold: DCoH-likeSuperfamily: C-terminal domain of arginine repressorFamily: C-terminal domain of arginine repressor66 c2pjkA_
not modelled
12.0
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 178aa long hypothetical molybdenum cofactorPDBTitle: structure of hypothetical molybdenum cofactor biosynthesis2 protein b from sulfolobus tokodaii
67 c3lo3E_
not modelled
11.7
11
PDB header: structure genomics, unknown functionChain: E: PDB Molecule: uncharacterized conserved protein;PDBTitle: the crystal structure of a conserved functionally unknown2 protein from colwellia psychrerythraea 34h.
68 d2g2ca1
not modelled
11.6
16
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like69 c2l69A_
not modelled
11.2
19
PDB header: de novo proteinChain: A: PDB Molecule: rossmann 2x3 fold protein;PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or28
70 c1kb0A_
not modelled
11.1
23
PDB header: oxidoreductaseChain: A: PDB Molecule: quinohemoprotein alcohol dehydrogenase;PDBTitle: crystal structure of quinohemoprotein alcohol dehydrogenase from2 comamonas testosteroni
71 c2p2sA_
not modelled
11.1
11
PDB header: oxidoreductaseChain: A: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of putative oxidoreductase (yp_050235.1) from2 erwinia carotovora atroseptica scri1043 at 1.25 a resolution
72 d2fug12
not modelled
10.9
17
Fold: Nqo1 FMN-binding domain-likeSuperfamily: Nqo1 FMN-binding domain-likeFamily: Nqo1 FMN-binding domain-like73 c3nt5B_
not modelled
10.8
16
PDB header: oxidoreductaseChain: B: PDB Molecule: inositol 2-dehydrogenase/d-chiro-inositol 3-dehydrogenase;PDBTitle: crystal structure of myo-inositol dehydrogenase from bacillus subtilis2 with bound cofactor and product inosose
74 d1wu2a3
not modelled
10.8
13
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like75 c1yiqA_
not modelled
10.1
16
PDB header: oxidoreductaseChain: A: PDB Molecule: quinohemoprotein alcohol dehydrogenase;PDBTitle: molecular cloning and structural analysis of2 quinohemoprotein alcohol dehydrogenase adhiig from3 pseudomonas putida hk5. compariison to the other4 quinohemoprotein alcohol dehydrogenase adhiib found in the5 same microorganism.
76 c2nvwB_
not modelled
9.8
9
PDB header: transcriptionChain: B: PDB Molecule: galactose/lactose metabolism regulatory proteinPDBTitle: crystal sctucture of transcriptional regulator gal80p from2 kluyveromymes lactis
77 c2fugA_
not modelled
9.8
17
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-quinone oxidoreductase chain 1;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
78 d1ydwa1
not modelled
9.7
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain79 d1u2ca2
not modelled
9.3
23
Fold: Dystroglycan, domain 2Superfamily: Dystroglycan, domain 2Family: Dystroglycan, domain 280 c2qguA_
not modelled
9.2
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: probable signal peptide protein;PDBTitle: three-dimensional structure of the phospholipid-binding protein from2 ralstonia solanacearum q8xv73_ralsq in complex with a phospholipid at3 the resolution 1.53 a. northeast structural genomics consortium4 target rsr89
81 c3gfgB_
not modelled
9.2
8
PDB header: oxidoreductaseChain: B: PDB Molecule: uncharacterized oxidoreductase yvaa;PDBTitle: structure of putative oxidoreductase yvaa from bacillus subtilis in2 triclinic form
82 c2q4eB_
not modelled
9.0
21
PDB header: oxidoreductaseChain: B: PDB Molecule: probable oxidoreductase at4g09670;PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at4g09670
83 c3ibpA_
not modelled
8.9
10
PDB header: cell cycleChain: A: PDB Molecule: chromosome partition protein mukb;PDBTitle: the crystal structure of the dimerization domain of escherichia coli2 structural maintenance of chromosomes protein mukb
84 d2ftsa3
not modelled
8.8
18
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like85 d2tpta3
not modelled
8.6
15
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain86 c1wx2A_
not modelled
8.5
15
PDB header: oxidoreductase/metal transportChain: A: PDB Molecule: tyrosinase;PDBTitle: crystal structure of the oxy-form of the copper-bound streptomyces2 castaneoglobisporus tyrosinase complexed with a caddie protein3 prepared by the addition of hydrogenperoxide
87 d2f7wa1
not modelled
8.3
14
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like88 d1srqa_
not modelled
8.2
11
Fold: Rap/Ran-GAPSuperfamily: Rap/Ran-GAPFamily: Rap/Ran-GAP89 c1kv9A_
not modelled
8.0
26
PDB header: oxidoreductaseChain: A: PDB Molecule: type ii quinohemoprotein alcohol dehydrogenase;PDBTitle: structure at 1.9 a resolution of a quinohemoprotein alcohol2 dehydrogenase from pseudomonas putida hk5
90 c1v9nA_
not modelled
7.9
19
PDB header: oxidoreductaseChain: A: PDB Molecule: malate dehydrogenase;PDBTitle: structure of malate dehydrogenase from pyrococcus horikoshii ot3
91 c2v9vA_
not modelled
7.9
9
PDB header: transcriptionChain: A: PDB Molecule: selenocysteine-specific elongation factor;PDBTitle: crystal structure of moorella thermoacetica selb(377-511)
92 d1xrha_
not modelled
7.8
16
Fold: L-sulfolactate dehydrogenase-likeSuperfamily: L-sulfolactate dehydrogenase-likeFamily: L-sulfolactate dehydrogenase-like93 c3uoeB_
not modelled
7.7
16
PDB header: oxidoreductaseChain: B: PDB Molecule: dehydrogenase;PDBTitle: the crystal structure of dehydrogenase from sinorhizobium meliloti
94 c2is8A_
not modelled
7.6
15
PDB header: structural proteinChain: A: PDB Molecule: molybdopterin biosynthesis enzyme, moab;PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
95 c2bonB_
not modelled
7.3
15
PDB header: transferaseChain: B: PDB Molecule: lipid kinase;PDBTitle: structure of an escherichia coli lipid kinase (yegs)
96 d2fiua1
not modelled
7.2
13
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Atu0297-like97 c3f4lF_
not modelled
7.2
13
PDB header: oxidoreductaseChain: F: PDB Molecule: putative oxidoreductase yhhx;PDBTitle: crystal structure of a probable oxidoreductase yhhx in2 triclinic form. northeast structural genomics target er647
98 d1yuaa2
not modelled
7.1
27
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Prokaryotic DNA topoisomerase I, a C-terminal fragment99 c3rfqC_
not modelled
7.1
24
PDB header: biosynthetic proteinChain: C: PDB Molecule: pterin-4-alpha-carbinolamine dehydratase moab2;PDBTitle: crystal structure of pterin-4-alpha-carbinolamine dehydratase moab22 from mycobacterium marinum