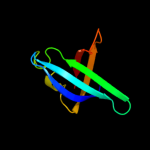
1 c3q8dB_
100.0
100
PDB header: dna binding proteinChain: B: PDB Molecule: dna repair protein reco;PDBTitle: e. coli reco complex with ssb c-terminus
2 c1u5kA_
100.0
17
PDB header: recombination,replicationChain: A: PDB Molecule: hypothetical protein;PDBTitle: recombinational repair protein reco
3 d1u5ka2
99.9
16
Fold: ArfGap/RecO-like zinc fingerSuperfamily: ArfGap/RecO-like zinc fingerFamily: RecO C-terminal domain-like4 d1u5ka1
98.4
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: RecO N-terminal domain-like5 d2dloa1
30.9
26
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain6 d1ibia2
27.3
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain7 d1rutx1
26.2
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain8 c2d9lA_
25.7
15
PDB header: gene regulationChain: A: PDB Molecule: nucleoporin-like protein rip;PDBTitle: solution structure of the arfgap domain of human rip
9 c2p57A_
24.3
13
PDB header: metal binding proteinChain: A: PDB Molecule: gtpase-activating protein znf289;PDBTitle: gap domain of znf289, an id1-regulated zinc finger protein
10 d1m3va1
23.7
21
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain11 c3iz5Z_
23.5
17
PDB header: ribosomeChain: Z: PDB Molecule: 60s ribosomal protein l24 (l24e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
12 c3izcZ_
22.5
8
PDB header: ribosomeChain: Z: PDB Molecule: 60s ribosomal protein rpl24 (l24e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
13 c4a1eT_
22.2
13
PDB header: ribosomeChain: T: PDB Molecule: rpl24;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
14 d1j2oa1
22.0
20
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain15 d1dcqa2
21.4
11
Fold: ArfGap/RecO-like zinc fingerSuperfamily: ArfGap/RecO-like zinc fingerFamily: Pyk2-associated protein beta ARF-GAP domain16 c2owaB_
21.1
8
PDB header: protein transportChain: B: PDB Molecule: arfgap-like finger domain containing protein;PDBTitle: crystal structure of putative gtpase activating protein for2 adp ribosylation factor from cryptosporidium parvum3 (cgd5_1040)
17 d1m4ma_
20.8
9
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat18 c3o47A_
20.3
11
PDB header: hydrolase, hydrolase activatorChain: A: PDB Molecule: adp-ribosylation factor gtpase-activating protein 1, adp-PDBTitle: crystal structure of arfgap1-arf1 fusion protein
19 c2iqjB_
19.8
16
PDB header: protein transportChain: B: PDB Molecule: stromal membrane-associated protein 1-like;PDBTitle: crystal structure of the gap domain of smap1l (loc64744)2 stromal membrane-associated protein 1-like
20 c2zkru_
19.5
26
PDB header: ribosomal protein/rnaChain: U: PDB Molecule: rna expansion segment es41;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
21 c3lvrE_
not modelled
19.4
13
PDB header: protein transportChain: E: PDB Molecule: arf-gap with sh3 domain, ank repeat and ph domain-PDBTitle: the crystal structure of asap3 in complex with arf6 in transition2 state soaked with calcium
22 d2ds5a1
not modelled
16.9
7
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: ClpX chaperone zinc binding domain23 c3mhsE_
not modelled
16.3
25
PDB header: hydrolase/transcription regulator/proteiChain: E: PDB Molecule: saga-associated factor 73;PDBTitle: structure of the saga ubp8/sgf11/sus1/sgf73 dub module bound to2 ubiquitin aldehyde
24 c1ovxB_
not modelled
16.3
7
PDB header: metal binding proteinChain: B: PDB Molecule: atp-dependent clp protease atp-binding subunit clpx;PDBTitle: nmr structure of the e. coli clpx chaperone zinc binding domain dimer
25 c3dwdB_
not modelled
15.6
11
PDB header: transport proteinChain: B: PDB Molecule: adp-ribosylation factor gtpase-activating protein 1;PDBTitle: crystal structure of the arfgap domain of human arfgap1
26 c1s1iS_
not modelled
15.0
9
PDB header: ribosomeChain: S: PDB Molecule: 60s ribosomal protein l24-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
27 d2cupa3
not modelled
14.7
15
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain28 c3k7aM_
not modelled
14.4
15
PDB header: transcriptionChain: M: PDB Molecule: transcription initiation factor iib;PDBTitle: crystal structure of an rna polymerase ii-tfiib complex
29 c2ds8A_
not modelled
14.0
8
PDB header: metal binding protein, protein bindingChain: A: PDB Molecule: atp-dependent clp protease atp-binding subunitPDBTitle: structure of the zbd-xb complex
30 c3na7A_
not modelled
13.9
12
PDB header: gene regulation, chaperoneChain: A: PDB Molecule: hp0958;PDBTitle: 2.2 angstrom structure of the hp0958 protein from helicobacter pylori2 ccug 17874
31 d1vqou1
not modelled
13.9
14
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Ribosomal protein L24e32 c3ccjU_
not modelled
13.9
14
PDB header: ribosomeChain: U: PDB Molecule: 50s ribosomal protein l24e;PDBTitle: structure of anisomycin resistant 50s ribosomal subunit: 23s rrna2 mutation c2534u
33 d2qfaa1
not modelled
13.6
9
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat34 c1x6aA_
not modelled
13.4
12
PDB header: protein bindingChain: A: PDB Molecule: lim domain kinase 2;PDBTitle: solution structures of the second lim domain of human lim-2 kinase 2 (limk2)
35 d1vd4a_
not modelled
13.0
9
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain36 d2raxa1
not modelled
12.9
9
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat37 d2cora1
not modelled
12.8
21
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain38 d1xb0b_
not modelled
12.4
23
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat39 c2b0oF_
not modelled
11.9
13
PDB header: metal binding proteinChain: F: PDB Molecule: uplc1;PDBTitle: crystal structure of uplc1 gap domain
40 c1dcqA_
not modelled
11.1
11
PDB header: metal binding proteinChain: A: PDB Molecule: pyk2-associated protein beta;PDBTitle: crystal structure of the arf-gap domain and ankyrin repeats2 of papbeta.
41 c2dloA_
not modelled
11.0
28
PDB header: cell adhesionChain: A: PDB Molecule: thyroid receptor-interacting protein 6;PDBTitle: solution structure of the second lim domain of human2 thyroid receptor-interacting protein 6
42 c3fehA_
not modelled
10.9
18
PDB header: hydrolase activatorChain: A: PDB Molecule: centaurin-alpha-1;PDBTitle: crystal structure of full length centaurin alpha-1
43 c2co8A_
not modelled
10.8
10
PDB header: metal binding proteinChain: A: PDB Molecule: nedd9 interacting protein with calponin homologyPDBTitle: solution structures of the lim domain of human nedd92 interacting protein with calponin homology and lim domains
44 d2qam01
not modelled
10.6
20
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L32p45 d1qbha_
not modelled
9.9
23
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat46 d1tfqa_
not modelled
9.9
27
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat47 d1q4qa_
not modelled
9.9
27
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat48 d1x3ha1
not modelled
9.8
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain49 d2j0151
not modelled
9.7
6
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L32p50 d2zjrz1
not modelled
9.6
19
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L32p51 c2kgoA_
not modelled
9.6
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ybii;PDBTitle: solution nmr structure of zn finger protein ybil from escherichia2 coli. nesg target et107, ocsp target ec0402
52 d2vsla1
not modelled
9.4
27
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat53 c3dzuD_
not modelled
8.9
29
PDB header: transcription/dnaChain: D: PDB Molecule: peroxisome proliferator-activated receptor gamma;PDBTitle: intact ppar gamma - rxr alpha nuclear receptor complex on dna bound2 with bvt.13, 9-cis retinoic acid and ncoa2 peptide
54 c1oy7C_
not modelled
8.9
14
PDB header: apoptosis/peptideChain: C: PDB Molecule: baculoviral iap repeat-containing protein 7;PDBTitle: structure and function analysis of peptide antagonists of melanoma2 inhibitor of apoptosis (ml-iap)
55 d2aqaa1
not modelled
8.8
17
Fold: Rubredoxin-likeSuperfamily: Nop10-like SnoRNPFamily: Nucleolar RNA-binding protein Nop10-like56 c2poiA_
not modelled
8.7
27
PDB header: signaling protein/apoptosisChain: A: PDB Molecule: baculoviral iap repeat-containing protein 4;PDBTitle: crystal structure of xiap bir1 domain (i222 form)
57 d1v4aa2
not modelled
8.6
14
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: GlnE-like domain58 d1u5sb1
not modelled
8.6
18
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain59 c2joxA_
not modelled
8.6
38
PDB header: transcriptionChain: A: PDB Molecule: churchill protein;PDBTitle: embryonic neural inducing factor churchill is not a dna-2 binding zinc finger protein: solution structure reveals a3 solvent-exposed beta-sheet and zinc binuclear cluster
60 c2k5cA_
not modelled
8.3
25
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein pf0385;PDBTitle: nmr structure for pf0385
61 c2vm5A_
not modelled
8.3
18
PDB header: apoptosisChain: A: PDB Molecule: baculoviral iap repeat-containing protein 1;PDBTitle: human bir2 domain of baculoviral inhibitor of apoptosis2 repeat-containing 1 (birc1)
62 c2p9lD_
not modelled
8.2
21
PDB header: structural proteinChain: D: PDB Molecule: actin-related protein 2/3 complex subunit 2;PDBTitle: crystal structure of bovine arp2/3 complex
63 c2kaeA_
not modelled
8.2
21
PDB header: transcription/dnaChain: A: PDB Molecule: gata-type transcription factor;PDBTitle: data-driven model of med1:dna complex
64 d1rutx3
not modelled
8.0
17
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain65 d2apob1
not modelled
8.0
18
Fold: Rubredoxin-likeSuperfamily: Nop10-like SnoRNPFamily: Nucleolar RNA-binding protein Nop10-like66 d1u5sb2
not modelled
7.9
30
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain67 c3m0aD_
not modelled
7.9
32
PDB header: signaling proteinChain: D: PDB Molecule: baculoviral iap repeat-containing protein 3;PDBTitle: crystal structure of traf2:ciap2 complex
68 d1osxa_
not modelled
7.9
25
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: BAFF receptor-like69 d1g73d_
not modelled
7.8
27
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat70 d1i3oe_
not modelled
7.7
27
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat71 d1jd5a_
not modelled
7.7
27
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat72 c1ibiA_
not modelled
7.6
22
PDB header: metal binding proteinChain: A: PDB Molecule: cysteine-rich protein 2;PDBTitle: quail cysteine and glycine-rich protein, nmr, 15 minimized2 model structures
73 c1x64A_
not modelled
7.4
12
PDB header: contractile proteinChain: A: PDB Molecule: alpha-actinin-2 associated lim protein;PDBTitle: solution structure of the lim domain of alpha-actinin-22 associated lim protein
74 d1k8kd2
not modelled
7.4
23
Fold: Secretion chaperone-likeSuperfamily: Arp2/3 complex subunitsFamily: Arp2/3 complex subunits75 c2gviA_
not modelled
7.2
16
PDB header: oxidoreductaseChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of a putative formylmethanofuran dehydrogenase2 subunit e (ta1109) from thermoplasma acidophilum at 1.87 a resolution
76 c1hk8A_
not modelled
7.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: anaerobic ribonucleotide-triphosphate reductase;PDBTitle: structural basis for allosteric substrate specificity2 regulation in class iii ribonucleotide reductases:3 nrdd in complex with dgtp
77 d1hk8a_
not modelled
7.2
15
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: Class III anaerobic ribonucleotide reductase NRDD subunit78 c1imlA_
not modelled
7.0
11
PDB header: metal-binding proteinChain: A: PDB Molecule: cysteine rich intestinal protein;PDBTitle: cysteine rich intestinal protein, nmr, 48 structures
79 d2ey4e1
not modelled
6.9
23
Fold: Rubredoxin-likeSuperfamily: Nop10-like SnoRNPFamily: Nucleolar RNA-binding protein Nop10-like80 c1p0tq_
not modelled
6.8
25
PDB header: protein bindingChain: Q: PDB Molecule: tumor necrosis factor receptor superfamilyPDBTitle: crystal structure of the baff-baff-r complex (part ii)
81 c3dwlI_
not modelled
6.7
31
PDB header: structural proteinChain: I: PDB Molecule: actin-related protein 2/3 complex subunit 2;PDBTitle: crystal structure of fission yeast arp2/3 complex lacking the arp22 subunit
82 c2kq9A_
not modelled
6.7
21
PDB header: transcriptionChain: A: PDB Molecule: dnak suppressor protein;PDBTitle: solution structure of dnak suppressor protein from2 agrobacterium tumefaciens c58. northeast structural3 genomics consortium target att12/ontario center for4 structural proteomics target atc0888
83 d1oqen_
not modelled
6.7
25
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: BAFF receptor-like84 d2dara2
not modelled
6.7
18
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain85 c1tjlD_
not modelled
6.7
23
PDB header: transcriptionChain: D: PDB Molecule: dnak suppressor protein;PDBTitle: crystal structure of transcription factor dksa from e. coli
86 d1xpaa2
not modelled
6.4
14
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: DNA repair factor XPA DNA- and RPA-binding domain, N-terminal subdomain87 c3jueA_
not modelled
6.4
10
PDB header: protein transport/endocytosisChain: A: PDB Molecule: arfgap with coiled-coil, ank repeat and ph domain-PDBTitle: crystal structure of arfgap and ank repeat domain of acap1
88 d1oqek_
not modelled
6.3
25
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: BAFF receptor-like89 d2dloa2
not modelled
6.3
15
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain90 d1te5a_
not modelled
6.3
8
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Class II glutamine amidotransferases91 d1wyha1
not modelled
6.2
24
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain92 c2eheA_
not modelled
6.1
9
PDB header: structural proteinChain: A: PDB Molecule: four and a half lim domains 3;PDBTitle: solution structure of the first lim domain from human four2 and a half lim domains protein 3
93 c3d00A_
not modelled
6.1
21
PDB header: metal binding proteinChain: A: PDB Molecule: tungsten formylmethanofuran dehydrogenase subunit e;PDBTitle: crystal structure of a tungsten formylmethanofuran dehydrogenase2 subunit e (fmde)-like protein (syn_00638) from syntrophus3 aciditrophicus at 1.90 a resolution
94 d1xb0a_
not modelled
6.0
24
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat95 c2yqpA_
not modelled
5.8
33
PDB header: gene regulation, hydrolaseChain: A: PDB Molecule: probable atp-dependent rna helicase ddx59;PDBTitle: solution structure of the zf-hit domain in dead (asp-glu-2 ala-asp) box polypeptide 59
96 c3hvzB_
not modelled
5.5
35
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the tgs domain of the clolep_03100 protein from2 clostridium leptum, northeast structural genomics consortium target3 qlr13a
97 d2dara1
not modelled
5.5
23
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain98 c2o13A_
not modelled
5.4
22
PDB header: metal binding proteinChain: A: PDB Molecule: muscle lim protein;PDBTitle: solution structure of the c-terminal lim domain of mlp/crp3
99 c1nnjA_
not modelled
5.4
17
PDB header: hydrolaseChain: A: PDB Molecule: formamidopyrimidine-dna glycosylase;PDBTitle: crystal structure complex between the lactococcus lactis fpg and an2 abasic site containing dna