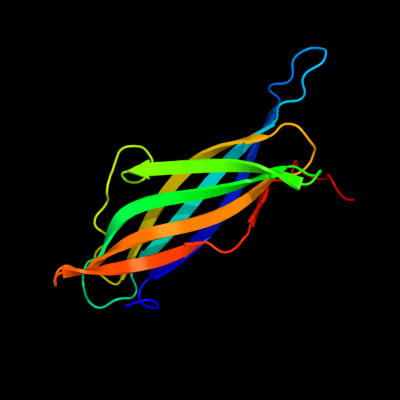
| 1 | d1tzaa_
|
|
 |
100.0 |
47 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:ApaG-like
Family:ApaG-like |
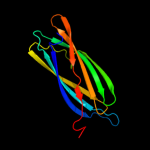
| 2 | d1xq4a_
|
|
 |
100.0 |
47 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:ApaG-like
Family:ApaG-like |
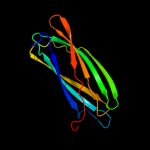
| 3 | d1xvsa_
|
|
 |
100.0 |
51 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:ApaG-like
Family:ApaG-like |
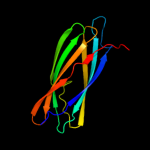
| 4 | c2f1eA_
|
|
 |
100.0 |
48 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein apag;
PDBTitle: solution structure of apag protein
|
| 5 | c3cfuA_
|
|
 |
81.2 |
13 |
PDB header:lipoprotein
Chain: A: PDB Molecule:uncharacterized lipoprotein yjha;
PDBTitle: crystal structure of the yjha protein from bacillus2 subtilis. northeast structural genomics consortium target3 sr562
|
| 6 | c3cg8B_
|
|
 |
79.2 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:laccase;
PDBTitle: laccase from streptomyces coelicolor
|
| 7 | c3cdzB_
|
|
 |
70.2 |
23 |
PDB header:blood clotting
Chain: B: PDB Molecule:coagulation factor viii light chain;
PDBTitle: crystal structure of human factor viii
|
| 8 | c2r7eB_
|
|
 |
67.6 |
23 |
PDB header:blood clotting
Chain: B: PDB Molecule:coagulation factor viii;
PDBTitle: crystal structure analysis of coagulation factor viii
|
| 9 | c2uxtA_
|
|
 |
64.5 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein sufi;
PDBTitle: sufi protein from escherichia coli
|
| 10 | c3rfrI_
|
|
 |
64.3 |
19 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:pmob;
PDBTitle: crystal structure of particulate methane monooxygenase (pmmo) from2 methylocystis sp. strain m
|
| 11 | d1cuoa_
|
|
 |
63.1 |
21 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 12 | c2x3bB_
|
|
 |
63.0 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:toxic extracellular endopeptidase;
PDBTitle: asap1 inactive mutant e294a, an extracellular toxic zinc2 metalloendopeptidase
|
| 13 | d1lm8v_
|
|
 |
60.0 |
25 |
Fold:Prealbumin-like
Superfamily:VHL
Family:VHL |
| 14 | c1sddA_
|
|
 |
59.9 |
17 |
PDB header:blood clotting
Chain: A: PDB Molecule:coagulation factor v;
PDBTitle: crystal structure of bovine factor vai
|
| 15 | d1hfua2
|
|
 |
59.5 |
21 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 16 | c2fqeA_
|
|
 |
57.8 |
36 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:blue copper oxidase cueo;
PDBTitle: crystal structures of e. coli laccase cueo under different2 copper binding situations
|
| 17 | c1l9mB_
|
|
 |
56.2 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:protein-glutamine glutamyltransferase e3;
PDBTitle: three-dimensional structure of the human transglutaminase 32 enzyme: binding of calcium ions change structure for3 activation
|
| 18 | d1nepa_
|
|
 |
55.1 |
15 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:ML domain |
| 19 | c3zx1A_
|
|
 |
53.8 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, putative;
PDBTitle: multicopper oxidase from campylobacter jejuni: a metallo-oxidase
|
| 20 | c3cdzA_
|
|
 |
52.8 |
15 |
PDB header:blood clotting
Chain: A: PDB Molecule:coagulation factor viii heavy chain;
PDBTitle: crystal structure of human factor viii
|
| 21 | c2xu9A_ |
|
not modelled |
51.3 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:laccase;
PDBTitle: crystal structure of laccase from thermus thermophilus hb27
|
| 22 | c3abgA_ |
|
not modelled |
50.4 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:bilirubin oxidase;
PDBTitle: x-ray crystal analysis of bilirubin oxidase from myrothecium2 verrucaria at 2.3 angstrom resolution using a twin crystal
|
| 23 | c2zwnA_ |
|
not modelled |
50.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:two-domain type laccase;
PDBTitle: crystal structure of the novel two-domain type laccase from a2 metagenome
|
| 24 | c2k6zA_ |
|
not modelled |
50.0 |
37 |
PDB header:metal transport
Chain: A: PDB Molecule:putative uncharacterized protein ttha1943;
PDBTitle: solution structures of copper loaded form pcua (trans2 conformation of the peptide bond involving the nitrogen of3 p14)
|
| 25 | c3m7oB_ |
|
not modelled |
48.7 |
23 |
PDB header:immune system
Chain: B: PDB Molecule:lymphocyte antigen 86;
PDBTitle: crystal structure of mouse md-1 in complex with phosphatidylcholine
|
| 26 | c3isyA_ |
|
not modelled |
46.9 |
16 |
PDB header:protein binding
Chain: A: PDB Molecule:intracellular proteinase inhibitor;
PDBTitle: crystal structure of an intracellular proteinase inhibitor (ipi,2 bsu11130) from bacillus subtilis at 2.61 a resolution
|
| 27 | d1azca_ |
|
not modelled |
46.5 |
24 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 28 | d2ccwa1 |
|
not modelled |
46.2 |
21 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 29 | d1v10a2 |
|
not modelled |
45.4 |
15 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 30 | c1yewI_ |
|
not modelled |
45.2 |
24 |
PDB header:oxidoreductase, membrane protein
Chain: I: PDB Molecule:particulate methane monooxygenase, b subunit;
PDBTitle: crystal structure of particulate methane monooxygenase
|
| 31 | c3kw8A_ |
|
not modelled |
42.7 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative copper oxidase;
PDBTitle: two-domain laccase from streptomyces coelicolor at 2.3 a resolution
|
| 32 | d1ibya_ |
|
not modelled |
41.6 |
15 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Nitrosocyanin |
| 33 | d1hc1a3 |
|
not modelled |
41.3 |
30 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Arthropod hemocyanin, C-terminal domain |
| 34 | c3nrfA_ |
|
not modelled |
41.2 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:apag protein;
PDBTitle: crystal structure of an apag protein (pa1934) from pseudomonas2 aeruginosa pao1 at 1.50 a resolution
|
| 35 | c1sddB_ |
|
not modelled |
40.0 |
28 |
PDB header:blood clotting
Chain: B: PDB Molecule:coagulation factor v;
PDBTitle: crystal structure of bovine factor vai
|
| 36 | c3g5wC_ |
|
not modelled |
39.1 |
23 |
PDB header:metal binding protein
Chain: C: PDB Molecule:multicopper oxidase type 1;
PDBTitle: crystal structure of blue copper oxidase from nitrosomonas europaea
|
| 37 | d1ufga_ |
|
not modelled |
38.1 |
17 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Lamin A/C globular tail domain
Family:Lamin A/C globular tail domain |
| 38 | d2plta_ |
|
not modelled |
37.4 |
22 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 39 | c3jt0B_ |
|
not modelled |
36.9 |
11 |
PDB header:structural protein
Chain: B: PDB Molecule:lamin-b1;
PDBTitle: crystal structure of the c-terminal fragment (426-558)2 lamin-b1 from homo sapiens, northeast structural genomics3 consortium target hr5546a
|
| 40 | c1v10A_ |
|
not modelled |
35.1 |
17 |
PDB header:oxidase
Chain: A: PDB Molecule:laccase;
PDBTitle: structure of rigidoporus lignosus laccase from hemihedrally2 twinned crystals
|
| 41 | d1cc3a_ |
|
not modelled |
34.8 |
25 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 42 | c2bnoA_ |
|
not modelled |
34.2 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:epoxidase;
PDBTitle: the structure of hydroxypropylphosphonic acid epoxidase2 from s. wedmorenis.
|
| 43 | d2j5wa3 |
|
not modelled |
34.0 |
32 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 44 | c3rgbA_ |
|
not modelled |
33.9 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methane monooxygenase subunit b2;
PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
|
| 45 | c2zooA_ |
|
not modelled |
33.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable nitrite reductase;
PDBTitle: crystal structure of nitrite reductase from pseudoalteromonas2 haloplanktis tac125
|
| 46 | c1wa1X_ |
|
not modelled |
33.1 |
17 |
PDB header:reductase
Chain: X: PDB Molecule:dissimilatory copper-containing nitrite
PDBTitle: crystal structure of h313q mutant of alcaligenes2 xylosoxidans nitrite reductase
|
| 47 | d1jzga_ |
|
not modelled |
31.6 |
21 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 48 | c2h47C_ |
|
not modelled |
30.9 |
24 |
PDB header:oxidoreductase/electron transport
Chain: C: PDB Molecule:azurin;
PDBTitle: crystal structure of an electron transfer complex between2 aromatic amine dephydrogenase and azurin from alcaligenes3 faecalis (form 1)
|
| 49 | d1gyca2 |
|
not modelled |
30.7 |
13 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 50 | d1ifra_ |
|
not modelled |
30.7 |
17 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Lamin A/C globular tail domain
Family:Lamin A/C globular tail domain |
| 51 | d1joia_ |
|
not modelled |
30.5 |
18 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 52 | d1ejxb_ |
|
not modelled |
29.7 |
31 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 53 | d4ubpb_ |
|
not modelled |
28.1 |
31 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 54 | d1e9ya1 |
|
not modelled |
27.0 |
33 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
| 55 | d1sdda1 |
|
not modelled |
27.0 |
17 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 56 | c3mu3A_ |
|
not modelled |
26.9 |
13 |
PDB header:immune system
Chain: A: PDB Molecule:protein md-1;
PDBTitle: crystal structure of chicken md-1 complexed with lipid iva
|
| 57 | d2j5wa1 |
|
not modelled |
25.7 |
17 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 58 | d1rkra_ |
|
not modelled |
24.9 |
19 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 59 | d1nwpa_ |
|
not modelled |
24.8 |
22 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 60 | d1kyaa2 |
|
not modelled |
23.9 |
17 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 61 | d2q9oa1 |
|
not modelled |
23.2 |
19 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 62 | d1hfua1 |
|
not modelled |
23.1 |
19 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 63 | d1rutx4 |
|
not modelled |
21.9 |
27 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 64 | c1mzzC_ |
|
not modelled |
21.6 |
37 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:copper-containing nitrite reductase;
PDBTitle: crystal structure of mutant (m182t)of nitrite reductase
|
| 65 | c3gdcC_ |
|
not modelled |
21.1 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:multicopper oxidase;
PDBTitle: crystal structure of multicopper oxidase
|
| 66 | d1qbaa2 |
|
not modelled |
21.0 |
15 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Carbohydrate-binding domain
Family:Bacterial chitobiase, n-terminal domain |
| 67 | d1kv7a2 |
|
not modelled |
20.7 |
16 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 68 | c1kcwA_ |
|
not modelled |
20.3 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ceruloplasmin;
PDBTitle: x-ray crystal structure of human ceruloplasmin at 3.0 angstroms
|
| 69 | d1ndsa1 |
|
not modelled |
20.2 |
17 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 70 | d1ivta_ |
|
not modelled |
20.1 |
17 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Lamin A/C globular tail domain
Family:Lamin A/C globular tail domain |
| 71 | c1e9zA_ |
|
not modelled |
19.9 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:urease subunit alpha;
PDBTitle: crystal structure of helicobacter pylori urease
|
| 72 | d1hcza2 |
|
not modelled |
19.8 |
22 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:Cytochrome f, small domain |
| 73 | d1kv7a1 |
|
not modelled |
19.8 |
26 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 74 | c3qgaD_ |
|
not modelled |
19.7 |
11 |
PDB header:hydrolase
Chain: D: PDB Molecule:fusion of urease beta and gamma subunits;
PDBTitle: 3.0 a model of iron containing urease urea2b2 from helicobacter2 mustelae
|
| 75 | d1aoza2 |
|
not modelled |
18.8 |
19 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 76 | d1sddb1 |
|
not modelled |
17.9 |
40 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 77 | d1oe1a1 |
|
not modelled |
17.4 |
14 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 78 | d1exha_ |
|
not modelled |
16.9 |
43 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Carbohydrate-binding domain
Family:Cellulose-binding domain family II |
| 79 | d1kbva2 |
|
not modelled |
16.8 |
14 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 80 | c2yxwB_ |
|
not modelled |
16.6 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:blue copper oxidase cueo;
PDBTitle: the deletion mutant of multicopper oxidase cueo
|
| 81 | d2q9oa2 |
|
not modelled |
16.6 |
15 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 82 | c3orjA_ |
|
not modelled |
16.3 |
20 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sugar-binding protein;
PDBTitle: crystal structure of a sugar-binding protein (bacova_04391) from2 bacteroides ovatus at 2.16 a resolution
|
| 83 | d1kyaa1 |
|
not modelled |
15.9 |
12 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 84 | c3h8uA_ |
|
not modelled |
14.7 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized conserved protein with double-stranded
PDBTitle: crystal structure of uncharacterized conserved protein with double-2 stranded beta-helix domain (yp_001338853.1) from klebsiella3 pneumoniae subsp. pneumoniae mgh 78578 at 1.80 a resolution
|
| 85 | d1v97a4 |
|
not modelled |
13.4 |
15 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:CO dehydrogenase flavoprotein C-terminal domain-like
Family:CO dehydrogenase flavoprotein C-terminal domain-like |
| 86 | c3hhsB_ |
|
not modelled |
13.3 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:phenoloxidase subunit 1;
PDBTitle: crystal structure of manduca sexta prophenoloxidase
|
| 87 | d1gyca1 |
|
not modelled |
13.2 |
13 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 88 | c1kbwA_ |
|
not modelled |
13.1 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:major outer membrane protein pan 1;
PDBTitle: crystal structure of the soluble domain of ania from2 neisseria gonorrhoeae
|
| 89 | d2ar1a1 |
|
not modelled |
13.0 |
17 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:Atu2648/PH1033-like |
| 90 | d1m1ha1 |
|
not modelled |
12.9 |
33 |
Fold:N-utilization substance G protein NusG, insert domain
Superfamily:N-utilization substance G protein NusG, insert domain
Family:N-utilization substance G protein NusG, insert domain |
| 91 | d1aoza1 |
|
not modelled |
12.4 |
19 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 92 | d2bnma2 |
|
not modelled |
12.2 |
11 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:TM1459-like |
| 93 | c1asqB_ |
|
not modelled |
11.8 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ascorbate oxidase;
PDBTitle: x-ray structures and mechanistic implications of three functional2 derivatives of ascorbate oxidase from zucchini: reduced-, peroxide-,3 and azide-forms
|
| 94 | d2fdbm1 |
|
not modelled |
11.6 |
13 |
Fold:beta-Trefoil
Superfamily:Cytokine
Family:Fibroblast growth factors (FGF) |
| 95 | d1llaa3 |
|
not modelled |
11.5 |
25 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Arthropod hemocyanin, C-terminal domain |
| 96 | c3ixvG_ |
|
not modelled |
10.6 |
25 |
PDB header:oxygen binding
Chain: G: PDB Molecule:hemocyanin aa6 chain;
PDBTitle: scorpion hemocyanin resting state pseudo atomic model built based on2 cryo-em density map
|
| 97 | c3d5bN_ |
|
not modelled |
10.5 |
14 |
PDB header:ribosome
Chain: N: PDB Molecule:50s ribosomal protein l13;
PDBTitle: structural basis for translation termination on the 70s ribosome. this2 file contains the 50s subunit of one 70s ribosome. the entire crystal3 structure contains two 70s ribosomes as described in remark 400.
|
| 98 | d2zjrg1 |
|
not modelled |
10.5 |
17 |
Fold:Ribosomal protein L13
Superfamily:Ribosomal protein L13
Family:Ribosomal protein L13 |
| 99 | c3cf5G_ |
|
not modelled |
10.5 |
17 |
PDB header:ribosome/antibiotic
Chain: G: PDB Molecule:50s ribosomal protein l13;
PDBTitle: thiopeptide antibiotic thiostrepton bound to the large ribosomal2 subunit of deinococcus radiodurans
|