| 1 | c2qlcC_ |
|
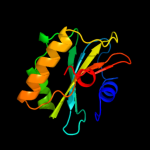 |
100.0 |
38 |
PDB header:dna binding protein
Chain: C: PDB Molecule:dna repair protein radc homolog;
PDBTitle: the crystal structure of dna repair protein radc from chlorobium2 tepidum tls
|
| 2 | d1oi0a_ |
|
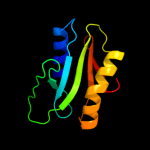 |
96.8 |
21 |
Fold:Cytidine deaminase-like
Superfamily:JAB1/MPN domain
Family:JAB1/MPN domain |
| 3 | c2kcqA_ |
|
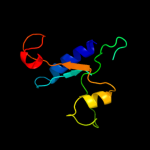 |
96.2 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:mov34/mpn/pad-1 family;
PDBTitle: solution structure of protein sru_2040 from salinibacter2 ruber (strain dsm 13855) . northeast structural genomics3 consortium target srr106
|
| 4 | c2kksA_ |
|
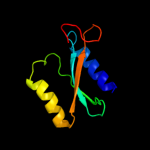 |
96.1 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of protein dsy2949 from desulfitobacterium2 hafniense. northeast structural genomics consortium target dhr27
|
| 5 | c2w6rA_ |
|
 |
82.3 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:imidazole glycerol phosphate synthase subunit
PDBTitle: crystal structure of an artificial (ba)8-barrel protein2 designed from identical half barrels
|
| 6 | c3tdmD_ |
|
 |
70.1 |
12 |
PDB header:de novo protein
Chain: D: PDB Molecule:computationally designed two-fold symmetric tim-barrel
PDBTitle: computationally designed tim-barrel protein, halfflr
|
| 7 | c2yciX_ |
|
 |
62.8 |
20 |
PDB header:transferase
Chain: X: PDB Molecule:5-methyltetrahydrofolate corrinoid/iron sulfur protein
PDBTitle: methyltransferase native
|
| 8 | d2csua1 |
|
 |
62.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 9 | c3jrkG_ |
|
 |
62.2 |
21 |
PDB header:lyase
Chain: G: PDB Molecule:tagatose 1,6-diphosphate aldolase 2;
PDBTitle: a putative tagatose 1,6-diphosphate aldolase from streptococcus2 pyogenes
|
| 10 | c3q94B_ |
|
 |
60.7 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase, class ii;
PDBTitle: the crystal structure of fructose 1,6-bisphosphate aldolase from2 bacillus anthracis str. 'ames ancestor'
|
| 11 | d1gvfa_ |
|
 |
59.2 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 12 | c3c52B_ |
|
 |
55.8 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: class ii fructose-1,6-bisphosphate aldolase from2 helicobacter pylori in complex with3 phosphoglycolohydroxamic acid, a competitive inhibitor
|
| 13 | c2iswB_ |
|
 |
54.6 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-1,6-bisphosphate aldolase;
PDBTitle: structure of giardia fructose-1,6-biphosphate aldolase in2 complex with phosphoglycolohydroxamate
|
| 14 | c3pm6B_ |
|
 |
54.6 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-bisphosphate aldolase;
PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
|
| 15 | d1rvga_ |
|
 |
51.7 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 16 | d1f6ya_ |
|
 |
51.4 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 17 | c3e49A_ |
|
 |
44.2 |
25 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849 with a tim barrel fold;
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 with a tim barrel fold (bxe_c0966) from burkholderia xenovorans lb4003 at 1.75 a resolution
|
| 18 | c3op1A_ |
|
 |
39.5 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:macrolide-efflux protein;
PDBTitle: crystal structure of macrolide-efflux protein sp_1110 from2 streptococcus pneumoniae
|
| 19 | c3elfA_ |
|
 |
38.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: structural characterization of tetrameric mycobacterium tuberculosis2 fructose 1,6-bisphosphate aldolase - substrate binding and catalysis3 mechanism of a class iia bacterial aldolase
|
| 20 | d1hl9a2 |
|
 |
37.5 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Putative alpha-L-fucosidase, catalytic domain |
| 21 | c3t18D_ |
|
not modelled |
32.9 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:aminotransferase class i and ii;
PDBTitle: crystal structure of aminotransferase from anaerococcus prevotii dsm2 20548.
|
| 22 | c3qm3C_ |
|
not modelled |
32.9 |
9 |
PDB header:lyase
Chain: C: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: 1.85 angstrom resolution crystal structure of fructose-bisphosphate2 aldolase (fba) from campylobacter jejuni
|
| 23 | d1dosa_ |
|
not modelled |
32.6 |
2 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 24 | c3stgA_ |
|
not modelled |
30.6 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of a58p, del(n59), and loop 7 truncated mutant of 3-2 deoxy-d-manno-octulosonate 8-phosphate synthase (kdo8ps) from3 neisseria meningitidis
|
| 25 | c2znrA_ |
|
not modelled |
29.8 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:amsh-like protease;
PDBTitle: crystal structure of the dub domain of human amsh-lp
|
| 26 | c3gndC_ |
|
not modelled |
28.6 |
18 |
PDB header:lyase
Chain: C: PDB Molecule:aldolase lsrf;
PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
|
| 27 | c3k13A_ |
|
not modelled |
28.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydrofolate-homocysteine methyltransferase;
PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
|
| 28 | d2isya2 |
|
not modelled |
28.0 |
25 |
Fold:Iron-dependent repressor protein, dimerization domain
Superfamily:Iron-dependent repressor protein, dimerization domain
Family:Iron-dependent repressor protein, dimerization domain |
| 29 | d3bzka5 |
|
not modelled |
27.0 |
15 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Tex RuvX-like domain-like |
| 30 | c2x0kB_ |
|
not modelled |
26.7 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin biosynthesis protein ribf;
PDBTitle: crystal structure of modular fad synthetase from2 corynebacterium ammoniagenes
|
| 31 | d1je0a_ |
|
not modelled |
24.2 |
17 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 32 | c3c6cA_ |
|
not modelled |
23.5 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of a putative 3-keto-5-aminohexanoate cleavage2 enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 a3 resolution
|
| 33 | d2axtu1 |
|
not modelled |
23.2 |
20 |
Fold:SAM domain-like
Superfamily:PsbU/PolX domain-like
Family:PsbU-like |
| 34 | c1s5lu_ |
|
not modelled |
21.7 |
20 |
PDB header:photosynthesis
Chain: U: PDB Molecule:photosystem ii 12 kda extrinsic protein;
PDBTitle: architecture of the photosynthetic oxygen evolving center
|
| 35 | c3lotC_ |
|
not modelled |
21.2 |
17 |
PDB header:structure genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein of unknown function (np_070038.1) from2 archaeoglobus fulgidus at 1.89 a resolution
|
| 36 | c3eypB_ |
|
not modelled |
20.9 |
23 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of putative alpha-l-fucosidase from bacteroides2 thetaiotaomicron
|
| 37 | d1ybfa_ |
|
not modelled |
20.5 |
8 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 38 | c2xrfA_ |
|
not modelled |
20.2 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:uridine phosphorylase 2;
PDBTitle: crystal structure of human uridine phosphorylase 2
|
| 39 | d1ka9f_ |
|
not modelled |
19.2 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 40 | c3fiuD_ |
|
not modelled |
18.5 |
16 |
PDB header:ligase
Chain: D: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: structure of nmn synthetase from francisella tularensis
|
| 41 | d2j9ga2 |
|
not modelled |
18.0 |
9 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 42 | c3mo4B_ |
|
not modelled |
17.7 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-1,3/4-fucosidase;
PDBTitle: the crystal structure of an alpha-(1-3,4)-fucosidase from2 bifidobacterium longum subsp. infantis atcc 15697
|
| 43 | c2y85D_ |
|
not modelled |
17.5 |
11 |
PDB header:isomerase
Chain: D: PDB Molecule:phosphoribosyl isomerase a;
PDBTitle: crystal structure of mycobacterium tuberculosis phosphoribosyl2 isomerase with bound rcdrp
|
| 44 | d1e0fi_ |
|
not modelled |
16.5 |
44 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Leech antihemostatic proteins
Family:Hirudin-like |
| 45 | c1e0fI_ |
|
not modelled |
16.5 |
44 |
PDB header:coagulation/crystal structure/heparin-b
Chain: I: PDB Molecule:haemadin;
PDBTitle: crystal structure of the human alpha-thrombin-haemadin2 complex: an exosite ii-binding inhibitor
|
| 46 | c1kftA_ |
|
not modelled |
16.1 |
18 |
PDB header:dna binding protein
Chain: A: PDB Molecule:excinuclease abc subunit c;
PDBTitle: solution structure of the c-terminal domain of uvrc from e-2 coli
|
| 47 | d1kfta_ |
|
not modelled |
16.1 |
18 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Excinuclease UvrC C-terminal domain |
| 48 | c3guzB_ |
|
not modelled |
15.9 |
11 |
PDB header:ligase
Chain: B: PDB Molecule:pantothenate synthetase;
PDBTitle: structural and substrate-binding studies of pantothenate2 synthenate (ps)provide insights into homotropic inhibition3 by pantoate in ps's
|
| 49 | c2bdqA_ |
|
not modelled |
15.6 |
17 |
PDB header:metal transport
Chain: A: PDB Molecule:copper homeostasis protein cutc;
PDBTitle: crystal structure of the putative copper homeostasis2 protein cutc from streptococcus agalactiae, northeast3 strucural genomics target sar15.
|
| 50 | c3iz5w_ |
|
not modelled |
15.0 |
50 |
PDB header:ribosome
Chain: W: PDB Molecule:60s ribosomal protein l22 (l22e);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 51 | d1oxja2 |
|
not modelled |
14.6 |
12 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:PHAT domain |
| 52 | c1e0fJ_ |
|
not modelled |
14.6 |
44 |
PDB header:coagulation/crystal structure/heparin-b
Chain: J: PDB Molecule:haemadin;
PDBTitle: crystal structure of the human alpha-thrombin-haemadin2 complex: an exosite ii-binding inhibitor
|
| 53 | d1xi3a_ |
|
not modelled |
14.5 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
| 54 | d1x2ia1 |
|
not modelled |
14.4 |
18 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Hef domain-like |
| 55 | d1ulza2 |
|
not modelled |
14.4 |
14 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 56 | d1pzna1 |
|
not modelled |
14.3 |
25 |
Fold:SAM domain-like
Superfamily:Rad51 N-terminal domain-like
Family:DNA repair protein Rad51, N-terminal domain |
| 57 | d1w5da1 |
|
not modelled |
14.2 |
16 |
Fold:beta-lactamase/transpeptidase-like
Superfamily:beta-lactamase/transpeptidase-like
Family:Dac-like |
| 58 | c2w1oA_ |
|
not modelled |
13.8 |
33 |
PDB header:translation
Chain: A: PDB Molecule:60s acidic ribosomal protein p2;
PDBTitle: nmr structure of dimerization domain of human ribosomal2 protein p2
|
| 59 | d1ywxa1 |
|
not modelled |
13.7 |
24 |
Fold:Ribosomal proteins S24e, L23 and L15e
Superfamily:Ribosomal proteins S24e, L23 and L15e
Family:Ribosomal protein S24e |
| 60 | d2ex2a1 |
|
not modelled |
13.7 |
16 |
Fold:beta-lactamase/transpeptidase-like
Superfamily:beta-lactamase/transpeptidase-like
Family:Dac-like |
| 61 | c1t3tA_ |
|
not modelled |
13.5 |
80 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine synthase;
PDBTitle: structure of formylglycinamide synthetase
|
| 62 | d1k9sa_ |
|
not modelled |
13.0 |
14 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 63 | d2a1jb1 |
|
not modelled |
12.8 |
32 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Hef domain-like |
| 64 | c1e0fK_ |
|
not modelled |
12.5 |
44 |
PDB header:coagulation/crystal structure/heparin-b
Chain: K: PDB Molecule:haemadin;
PDBTitle: crystal structure of the human alpha-thrombin-haemadin2 complex: an exosite ii-binding inhibitor
|
| 65 | d1ii7a_ |
|
not modelled |
12.4 |
20 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DNA double-strand break repair nuclease |
| 66 | c1jvnB_ |
|
not modelled |
12.4 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional histidine biosynthesis protein hishf;
PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
|
| 67 | c3av0A_ |
|
not modelled |
12.3 |
20 |
PDB header:recombination
Chain: A: PDB Molecule:dna double-strand break repair protein mre11;
PDBTitle: crystal structure of mre11-rad50 bound to atp s
|
| 68 | d1h5ya_ |
|
not modelled |
12.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 69 | d2i1qa1 |
|
not modelled |
12.2 |
15 |
Fold:SAM domain-like
Superfamily:Rad51 N-terminal domain-like
Family:DNA repair protein Rad51, N-terminal domain |
| 70 | d1g3wa2 |
|
not modelled |
11.9 |
25 |
Fold:Iron-dependent repressor protein, dimerization domain
Superfamily:Iron-dependent repressor protein, dimerization domain
Family:Iron-dependent repressor protein, dimerization domain |
| 71 | d2aq0a1 |
|
not modelled |
11.8 |
11 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Hef domain-like |
| 72 | c2j8qB_ |
|
not modelled |
11.8 |
20 |
PDB header:nuclear protein
Chain: B: PDB Molecule:cleavage and polyadenylation specificity factor 5;
PDBTitle: crystal structure of human cleavage and polyadenylation2 specificity factor 5 (cpsf5) in complex with a sulphate3 ion.
|
| 73 | c3iwpK_ |
|
not modelled |
11.8 |
15 |
PDB header:metal binding protein
Chain: K: PDB Molecule:copper homeostasis protein cutc homolog;
PDBTitle: crystal structure of human copper homeostasis protein cutc
|
| 74 | c2csuB_ |
|
not modelled |
11.7 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:457aa long hypothetical protein;
PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
|
| 75 | d1t3ta4 |
|
not modelled |
11.5 |
67 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
| 76 | c2it0A_ |
|
not modelled |
11.4 |
25 |
PDB header:transcription/dna
Chain: A: PDB Molecule:iron-dependent repressor ider;
PDBTitle: crystal structure of a two-domain ider-dna complex crystal2 form ii
|
| 77 | d1s7ia_ |
|
not modelled |
11.3 |
6 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:DGPF domain (Pfam 04946) |
| 78 | c3a3eB_ |
|
not modelled |
11.2 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:penicillin-binding protein 4;
PDBTitle: crystal structure of penicillin binding protein 4 (dacb)2 from haemophilus influenzae, complexed with novel beta-3 lactam (cmv)
|
| 79 | d1xn9a_ |
|
not modelled |
11.1 |
20 |
Fold:Ribosomal proteins S24e, L23 and L15e
Superfamily:Ribosomal proteins S24e, L23 and L15e
Family:Ribosomal protein S24e |
| 80 | c3no5C_ |
|
not modelled |
10.8 |
15 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a pfam duf849 domain containing protein2 (reut_a1631) from ralstonia eutropha jmp134 at 1.90 a resolution
|
| 81 | c2xzmP_ |
|
not modelled |
10.7 |
28 |
PDB header:ribosome
Chain: P: PDB Molecule:rps24e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
|
| 82 | c2deoA_ |
|
not modelled |
10.7 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:441aa long hypothetical nfed protein;
PDBTitle: 1510-n membrane protease specific for a stomatin homolog from2 pyrococcus horikoshii
|
| 83 | c2x5fB_ |
|
not modelled |
10.6 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:aspartate_tyrosine_phenylalanine pyridoxal-5'
PDBTitle: crystal structure of the methicillin-resistant2 staphylococcus aureus sar2028, an3 aspartate_tyrosine_phenylalanine pyridoxal-5'-phosphate4 dependent aminotransferase
|
| 84 | d2qi2a3 |
|
not modelled |
10.5 |
15 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:ERF1/Dom34 C-terminal domain-like |
| 85 | c3chvA_ |
|
not modelled |
10.4 |
16 |
PDB header:metal binding protein
Chain: A: PDB Molecule:prokaryotic domain of unknown function (duf849) with a tim
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
|
| 86 | c3iz6U_ |
|
not modelled |
10.2 |
40 |
PDB header:ribosome
Chain: U: PDB Molecule:40s ribosomal protein s24 (s24e);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 87 | c3izbU_ |
|
not modelled |
10.2 |
9 |
PDB header:ribosome
Chain: U: PDB Molecule:40s ribosomal protein s24;
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 88 | c2aw5A_ |
|
not modelled |
10.1 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp-dependent malic enzyme;
PDBTitle: crystal structure of a human malic enzyme
|
| 89 | c2hnhA_ |
|
not modelled |
10.0 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase iii alpha subunit;
PDBTitle: crystal structure of the catalytic alpha subunit of e. coli2 replicative dna polymerase iii
|
| 90 | c3glvB_ |
|
not modelled |
10.0 |
11 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:lipopolysaccharide core biosynthesis protein;
PDBTitle: crystal structure of the lipopolysaccharide core biosynthesis protein2 from thermoplasma volcanium gss1
|
| 91 | c3e02A_ |
|
not modelled |
9.7 |
26 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849;
PDBTitle: crystal structure of a duf849 family protein (bxe_c0271) from2 burkholderia xenovorans lb400 at 1.90 a resolution
|
| 92 | c3zqoK_ |
|
not modelled |
9.6 |
17 |
PDB header:dna-binding protein
Chain: K: PDB Molecule:terminase small subunit;
PDBTitle: crystal structure of the small terminase oligomerization2 core domain from a spp1-like bacteriophage (crystal form 3)
|
| 93 | d1szpa1 |
|
not modelled |
9.4 |
25 |
Fold:SAM domain-like
Superfamily:Rad51 N-terminal domain-like
Family:DNA repair protein Rad51, N-terminal domain |
| 94 | c3kinB_ |
|
not modelled |
9.2 |
20 |
PDB header:motor protein
Chain: B: PDB Molecule:kinesin heavy chain;
PDBTitle: kinesin (dimeric) from rattus norvegicus
|
| 95 | d1szpb1 |
|
not modelled |
9.1 |
25 |
Fold:SAM domain-like
Superfamily:Rad51 N-terminal domain-like
Family:DNA repair protein Rad51, N-terminal domain |
| 96 | c2y7eA_ |
|
not modelled |
8.9 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of the 3-keto-5-aminohexanoate cleavage enzyme2 (kce) from candidatus cloacamonas acidaminovorans (tetragonal form)
|
| 97 | d1dfoa_ |
|
not modelled |
8.7 |
5 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 98 | c3f0hA_ |
|
not modelled |
8.4 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase;
PDBTitle: crystal structure of aminotransferase (rer070207000802) from2 eubacterium rectale at 1.70 a resolution
|
| 99 | c2wvsD_ |
|
not modelled |
8.3 |
23 |
PDB header:hydrolase
Chain: D: PDB Molecule:alpha-l-fucosidase;
PDBTitle: crystal structure of an alpha-l-fucosidase gh29 trapped2 covalent intermediate from bacteroides thetaiotaomicron in3 complex with 2-fluoro-fucosyl fluoride using an e288q4 mutant
|

