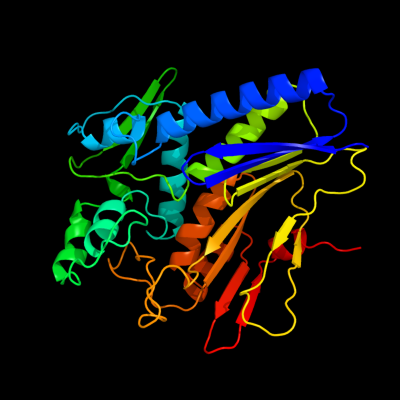
| 1 | c2o18A_
|
|
 |
100.0 |
97 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:thiamine biosynthesis lipoprotein apbe;
PDBTitle: crystal structure of a thiamine biosynthesis lipoprotein2 apbe, northeast strcutural genomics target er559
|
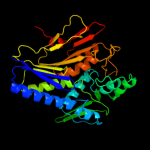
| 2 | d1vrma1
|
|
 |
100.0 |
26 |
Fold:T-fold
Superfamily:ApbE-like
Family:ApbE-like |
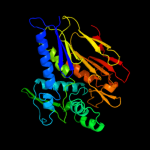
| 3 | d2o34a1
|
|
 |
98.5 |
21 |
Fold:T-fold
Superfamily:ApbE-like
Family:DVU1097-like |
| 4 | c2gpzC_
|
|
 |
35.7 |
42 |
PDB header:hydrolase
Chain: C: PDB Molecule:transthyretin-like protein;
PDBTitle: transthyretin-like protein from salmonella dublin
|
| 5 | c3qvaB_
|
|
 |
30.9 |
42 |
PDB header:hydrolase
Chain: B: PDB Molecule:transthyretin-like protein;
PDBTitle: structure of klebsiella pneumoniae 5-hydroxyisourate hydrolase
|
| 6 | d1udxa3
|
|
 |
30.4 |
30 |
Fold:Obg GTP-binding protein C-terminal domain
Superfamily:Obg GTP-binding protein C-terminal domain
Family:Obg GTP-binding protein C-terminal domain |
| 7 | d1f86a_
|
|
 |
27.9 |
42 |
Fold:Prealbumin-like
Superfamily:Transthyretin (synonym: prealbumin)
Family:Transthyretin (synonym: prealbumin) |
| 8 | d1kgia_
|
|
 |
27.6 |
37 |
Fold:Prealbumin-like
Superfamily:Transthyretin (synonym: prealbumin)
Family:Transthyretin (synonym: prealbumin) |
| 9 | d1tfpa_
|
|
 |
26.5 |
42 |
Fold:Prealbumin-like
Superfamily:Transthyretin (synonym: prealbumin)
Family:Transthyretin (synonym: prealbumin) |
| 10 | c2h0eA_
|
|
 |
26.1 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:transthyretin-like protein pucm;
PDBTitle: crystal structure of pucm in the absence of substrate
|
| 11 | c2h1xB_
|
|
 |
25.9 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:5-hydroxyisourate hydrolase (formerly known as
PDBTitle: crystal structure of 5-hydroxyisourate hydrolase (formerly2 known as trp, transthyretin related protein)
|
| 12 | d2vo1a1
|
|
 |
25.0 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 13 | d1ttaa_
|
|
 |
21.8 |
42 |
Fold:Prealbumin-like
Superfamily:Transthyretin (synonym: prealbumin)
Family:Transthyretin (synonym: prealbumin) |
| 14 | c3gehA_
|
|
 |
20.8 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:trna modification gtpase mnme;
PDBTitle: crystal structure of mnme from nostoc in complex with gdp, folinic2 acid and zn
|
| 15 | d1s1ma2
|
|
 |
20.8 |
33 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 16 | d1vcoa2
|
|
 |
20.3 |
22 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 17 | c2xw7A_
|
|
 |
17.3 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrofolate reductase;
PDBTitle: structure of mycobacterium smegmatis putative reductase ms0308
|
| 18 | d1t95a1
|
|
 |
16.5 |
50 |
Fold:RuvA C-terminal domain-like
Superfamily:Hypothetical protein AF0491, middle domain
Family:Hypothetical protein AF0491, middle domain |
| 19 | c2k10A_
|
|
 |
16.0 |
30 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:ranatuerin-2csa;
PDBTitle: confirmational analysis of the broad-spectrum antibacterial2 peptide, rantuerin-2csa: identification of a full length3 helix-turn-helix motif
|
| 20 | c3nvaB_
|
|
 |
14.9 |
30 |
PDB header:ligase
Chain: B: PDB Molecule:ctp synthase;
PDBTitle: dimeric form of ctp synthase from sulfolobus solfataricus
|
| 21 | c2o7pA_ |
|
not modelled |
14.4 |
21 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:riboflavin biosynthesis protein ribd;
PDBTitle: the crystal structure of ribd from escherichia coli in complex with2 the oxidised nadp+ cofactor in the active site of the reductase3 domain
|
| 22 | c2ad5B_ |
|
not modelled |
14.3 |
33 |
PDB header:ligase
Chain: B: PDB Molecule:ctp synthase;
PDBTitle: mechanisms of feedback regulation and drug resistance of ctp2 synthetases: structure of the e. coli ctps/ctp complex at 2.8-3 angstrom resolution.
|
| 23 | d2azna1 |
|
not modelled |
14.2 |
29 |
Fold:Dihydrofolate reductase-like
Superfamily:Dihydrofolate reductase-like
Family:RibD C-terminal domain-like |
| 24 | c3l51B_ |
|
not modelled |
14.0 |
15 |
PDB header:cell cycle
Chain: B: PDB Molecule:structural maintenance of chromosomes protein 4;
PDBTitle: crystal structure of the mouse condensin hinge domain
|
| 25 | c1vcnA_ |
|
not modelled |
13.9 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:ctp synthetase;
PDBTitle: crystal structure of t.th. hb8 ctp synthetase complex with sulfate2 anion
|
| 26 | c2d5nB_ |
|
not modelled |
13.7 |
25 |
PDB header:hydrolase, oxidoreductase
Chain: B: PDB Molecule:riboflavin biosynthesis protein ribd;
PDBTitle: crystal structure of a bifunctional deaminase and reductase2 involved in riboflavin biosynthesis
|
| 27 | d2gxfa1 |
|
not modelled |
13.6 |
19 |
Fold:Cystatin-like
Superfamily:NTF2-like
Family:YybH-like |
| 28 | d1gxja_ |
|
not modelled |
13.3 |
15 |
Fold:Smc hinge domain
Superfamily:Smc hinge domain
Family:Smc hinge domain |
| 29 | c2jppB_ |
|
not modelled |
13.3 |
21 |
PDB header:translation/rna
Chain: B: PDB Molecule:translational repressor;
PDBTitle: structural basis of rsma/csra rna recognition: structure of2 rsme bound to the shine-dalgarno sequence of hcna mrna
|
| 30 | d1v97a5 |
|
not modelled |
13.2 |
18 |
Fold:Molybdenum cofactor-binding domain
Superfamily:Molybdenum cofactor-binding domain
Family:Molybdenum cofactor-binding domain |
| 31 | c1vraA_ |
|
not modelled |
12.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:arginine biosynthesis bifunctional protein argj;
PDBTitle: crystal structure of arginine biosynthesis bifunctional protein argj2 (10175521) from bacillus halodurans at 2.00 a resolution
|
| 32 | d1cm3a_ |
|
not modelled |
12.3 |
31 |
Fold:HPr-like
Superfamily:HPr-like
Family:HPr-like |
| 33 | d2cyga1 |
|
not modelled |
12.2 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 34 | d2afhe1 |
|
not modelled |
11.6 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 35 | d1ffvb2 |
|
not modelled |
11.0 |
15 |
Fold:Molybdenum cofactor-binding domain
Superfamily:Molybdenum cofactor-binding domain
Family:Molybdenum cofactor-binding domain |
| 36 | d1g3qa_ |
|
not modelled |
10.8 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 37 | c2hxvA_ |
|
not modelled |
10.5 |
21 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:diaminohydroxyphosphoribosylaminopyrimidine deaminase/ 5-
PDBTitle: crystal structure of a diaminohydroxyphosphoribosylaminopyrimidine2 deaminase/ 5-amino-6-(5-phosphoribosylamino)uracil reductase (tm1828)3 from thermotoga maritima at 1.80 a resolution
|
| 38 | c2an1D_ |
|
not modelled |
10.4 |
30 |
PDB header:transferase
Chain: D: PDB Molecule:putative kinase;
PDBTitle: structural genomics, the crystal structure of a putative kinase from2 salmonella typhimurim lt2
|
| 39 | c1vpzB_ |
|
not modelled |
10.1 |
18 |
PDB header:rna binding protein
Chain: B: PDB Molecule:carbon storage regulator homolog;
PDBTitle: crystal structure of a putative carbon storage regulator protein2 (csra, pa0905) from pseudomonas aeruginosa at 2.05 a resolution
|
| 40 | d1vpza_ |
|
not modelled |
10.0 |
18 |
Fold:CsrA-like
Superfamily:CsrA-like
Family:CsrA-like |
| 41 | d2di9a1 |
|
not modelled |
9.7 |
20 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Filamin repeat (rod domain) |
| 42 | c3mwdA_ |
|
not modelled |
9.7 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:atp-citrate synthase;
PDBTitle: truncated human atp-citrate lyase with citrate bound
|
| 43 | d2b3za1 |
|
not modelled |
9.7 |
25 |
Fold:Dihydrofolate reductase-like
Superfamily:Dihydrofolate reductase-like
Family:RibD C-terminal domain-like |
| 44 | c3l51A_ |
|
not modelled |
9.4 |
20 |
PDB header:cell cycle
Chain: A: PDB Molecule:structural maintenance of chromosomes protein 2;
PDBTitle: crystal structure of the mouse condensin hinge domain
|
| 45 | c2qwuB_ |
|
not modelled |
9.4 |
14 |
PDB header:cell invasion
Chain: B: PDB Molecule:intracellular growth locus, subunit c;
PDBTitle: crystal structure of f. tularensis pathogenicity island2 protein c
|
| 46 | c2bekB_ |
|
not modelled |
9.4 |
29 |
PDB header:chromosome segregation
Chain: B: PDB Molecule:segregation protein;
PDBTitle: structure of the bacterial chromosome segregation protein2 soj
|
| 47 | d1a9xa3 |
|
not modelled |
9.4 |
19 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 48 | c2p4gA_ |
|
not modelled |
9.3 |
5 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a pyrimidine reductase-like protein (dip1392)2 from corynebacterium diphtheriae nctc at 2.30 a resolution
|
| 49 | d1n62b2 |
|
not modelled |
8.8 |
16 |
Fold:Molybdenum cofactor-binding domain
Superfamily:Molybdenum cofactor-binding domain
Family:Molybdenum cofactor-binding domain |
| 50 | c2kdoA_ |
|
not modelled |
8.8 |
40 |
PDB header:rna binding protein
Chain: A: PDB Molecule:ribosome maturation protein sbds;
PDBTitle: structure of the human shwachman-bodian-diamond syndrome protein, sbds
|
| 51 | c2vy8A_ |
|
not modelled |
8.6 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:polymerase basic protein 2;
PDBTitle: the 627-domain from influenza a virus polymerase pb22 subunit with glu-627
|
| 52 | c1t95A_ |
|
not modelled |
8.5 |
50 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein af0491;
PDBTitle: crystal structure of the shwachman-bodian-diamond syndrome2 protein orthologue from archaeoglobus fulgidus
|
| 53 | d1aq0a_ |
|
not modelled |
8.4 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 54 | d1zxoa1 |
|
not modelled |
8.2 |
19 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:BadF/BadG/BcrA/BcrD-like |
| 55 | d1zbsa2 |
|
not modelled |
7.9 |
23 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:BadF/BadG/BcrA/BcrD-like |
| 56 | d1oo2a_ |
|
not modelled |
7.8 |
16 |
Fold:Prealbumin-like
Superfamily:Transthyretin (synonym: prealbumin)
Family:Transthyretin (synonym: prealbumin) |
| 57 | c2panF_ |
|
not modelled |
7.7 |
26 |
PDB header:lyase
Chain: F: PDB Molecule:glyoxylate carboligase;
PDBTitle: crystal structure of e. coli glyoxylate carboligase
|
| 58 | c3iswA_ |
|
not modelled |
7.6 |
14 |
PDB header:structural protein
Chain: A: PDB Molecule:filamin-a;
PDBTitle: crystal structure of filamin-a immunoglobulin-like repeat 21 bound to2 an n-terminal peptide of cftr
|
| 59 | d1cp2a_ |
|
not modelled |
7.4 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 60 | d1g5ha2 |
|
not modelled |
7.3 |
8 |
Fold:Class II aaRS and biotin synthetases
Superfamily:Class II aaRS and biotin synthetases
Family:Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain |
| 61 | d1ghsa_ |
|
not modelled |
7.3 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 62 | c3htnA_ |
|
not modelled |
7.3 |
14 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative dna binding protein;
PDBTitle: crystal structure of a putative dna binding protein (bt_1116) from2 bacteroides thetaiotaomicron vpi-5482 at 1.50 a resolution
|
| 63 | d1i78a_ |
|
not modelled |
7.3 |
11 |
Fold:Transmembrane beta-barrels
Superfamily:OMPT-like
Family:Outer membrane protease OMPT |
| 64 | c2ojlB_ |
|
not modelled |
7.3 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of q7waf1_borpa from bordetella parapertussis.2 northeast structural genomics target bpr68.
|
| 65 | c3endA_ |
|
not modelled |
7.2 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:light-independent protochlorophyllide reductase
PDBTitle: crystal structure of the l protein of rhodobacter2 sphaeroides light-independent protochlorophyllide3 reductase (bchl) with mgadp bound: a homologue of the4 nitrogenase fe protein
|
| 66 | c3f55A_ |
|
not modelled |
7.1 |
10 |
PDB header:hydrolase, allergen
Chain: A: PDB Molecule:beta-1,3-glucanase;
PDBTitle: crystal structure of the native endo beta-1,3-glucanase (hev b 2), a2 major allergen from hevea brasiliensis (space group p41)
|
| 67 | c2obkE_ |
|
not modelled |
7.1 |
21 |
PDB header:structural genomics, unknown function
Chain: E: PDB Molecule:selt/selw/selh selenoprotein domain;
PDBTitle: x-ray structure of the putative se binding protein from pseudomonas2 fluorescens. northeast structural genomics consortium target plr6.
|
| 68 | d1g2914 |
|
not modelled |
7.0 |
44 |
Fold:OB-fold
Superfamily:MOP-like
Family:ABC-transporter additional domain |
| 69 | d1dvka_ |
|
not modelled |
7.0 |
9 |
Fold:Functional domain of the splicing factor Prp18
Superfamily:Functional domain of the splicing factor Prp18
Family:Functional domain of the splicing factor Prp18 |
| 70 | c2rc9A_ |
|
not modelled |
7.0 |
25 |
PDB header:membrane protein
Chain: A: PDB Molecule:glutamate [nmda] receptor subunit 3a;
PDBTitle: crystal structure of the nr3a ligand binding core complex with acpc at2 1.96 angstrom resolution
|
| 71 | d1ttea1 |
|
not modelled |
6.9 |
16 |
Fold:RuvA C-terminal domain-like
Superfamily:UBA-like
Family:UBA domain |
| 72 | c2wd5A_ |
|
not modelled |
6.9 |
15 |
PDB header:cell cycle
Chain: A: PDB Molecule:structural maintenance of chromosomes protein 1a;
PDBTitle: smc hinge heterodimer (mouse)
|
| 73 | d3e9la1 |
|
not modelled |
6.9 |
38 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Prp8 beta-finger domain-like |
| 74 | c2f7fA_ |
|
not modelled |
6.8 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase, putative;
PDBTitle: crystal structure of enterococcus faecalis putative nicotinate2 phosphoribosyltransferase, new york structural genomics consortium
|
| 75 | c2p0gB_ |
|
not modelled |
6.8 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:selenoprotein w-related protein;
PDBTitle: crystal structure of selenoprotein w-related protein from2 vibrio cholerae. northeast structural genomics target vcr75
|
| 76 | c3dexA_ |
|
not modelled |
6.8 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:sav_2001;
PDBTitle: crystal structure of sav_2001 protein from streptomyces2 avermitilis, northeast structural genomics consortium3 target svr107.
|
| 77 | d2dcfa1 |
|
not modelled |
6.7 |
3 |
Fold:beta-lactamase/transpeptidase-like
Superfamily:beta-lactamase/transpeptidase-like
Family:beta-Lactamase/D-ala carboxypeptidase |
| 78 | c2jtqA_ |
|
not modelled |
6.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phage shock protein e;
PDBTitle: rhodanese from e.coli
|
| 79 | c2fs1A_ |
|
not modelled |
6.7 |
24 |
PDB header:protein binding
Chain: A: PDB Molecule:psd-1;
PDBTitle: solution structure of psd-1
|
| 80 | d3e9oa1 |
|
not modelled |
6.7 |
38 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Prp8 beta-finger domain-like |
| 81 | c1t3oA_ |
|
not modelled |
6.7 |
18 |
PDB header:rna binding protein
Chain: A: PDB Molecule:carbon storage regulator;
PDBTitle: solution structure of csra, a bacterial carbon storage2 regulatory protein
|
| 82 | c3e66B_ |
|
not modelled |
6.5 |
38 |
PDB header:splicing
Chain: B: PDB Molecule:prp8;
PDBTitle: crystal structure of the beta-finger domain of yeast prp8
|
| 83 | d2i14a1 |
|
not modelled |
6.5 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 84 | d1g8fa3 |
|
not modelled |
6.4 |
10 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:ATP sulfurylase C-terminal domain |
| 85 | d1tifa_ |
|
not modelled |
6.3 |
15 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Translation initiation factor IF3, N-terminal domain
Family:Translation initiation factor IF3, N-terminal domain |
| 86 | d1p42a1 |
|
not modelled |
6.2 |
18 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC |
| 87 | c3kjgB_ |
|
not modelled |
6.1 |
25 |
PDB header:hydrolase, metal binding protein
Chain: B: PDB Molecule:co dehydrogenase/acetyl-coa synthase complex, accessory
PDBTitle: adp-bound state of cooc1
|
| 88 | d3enba1 |
|
not modelled |
6.1 |
38 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Prp8 beta-finger domain-like |
| 89 | c3jtwB_ |
|
not modelled |
6.1 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrofolate reductase;
PDBTitle: crystal structure of putative dihydrofolate reductase (yp_805003.1)2 from pediococcus pentosaceus atcc 25745 at 1.90 a resolution
|
| 90 | c2jyaA_ |
|
not modelled |
6.0 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu1810;
PDBTitle: nmr solution structure of protein atu1810 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr23,3 ontario centre for structural proteomics target atc1776
|
| 91 | d2isba1 |
|
not modelled |
6.0 |
19 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:FumA C-terminal domain-like
Family:FumA C-terminal domain-like |
| 92 | c3fg8B_ |
|
not modelled |
5.9 |
7 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein rha05790;
PDBTitle: crystal structure of pas domain of rha05790
|
| 93 | d1t3qb2 |
|
not modelled |
5.9 |
11 |
Fold:Molybdenum cofactor-binding domain
Superfamily:Molybdenum cofactor-binding domain
Family:Molybdenum cofactor-binding domain |
| 94 | c2lc2A_ |
|
not modelled |
5.8 |
14 |
PDB header:protein binding
Chain: A: PDB Molecule:avr3a4;
PDBTitle: solution structure of the rxlr effector p. capsici avr3a4
|
| 95 | c2nqcA_ |
|
not modelled |
5.8 |
23 |
PDB header:immune system
Chain: A: PDB Molecule:filamin-c;
PDBTitle: crystal structure of ig-like domain 23 from human filamin c
|
| 96 | d2nqca1 |
|
not modelled |
5.8 |
23 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Filamin repeat (rod domain) |
| 97 | d1a9xa4 |
|
not modelled |
5.7 |
19 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 98 | d2fa8a1 |
|
not modelled |
5.7 |
21 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Selenoprotein W-related |
| 99 | c3kgyA_ |
|
not modelled |
5.7 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:bifunctional deaminase-reductase domain protein;
PDBTitle: crystal structure of putative dihydrofolate reductase (yp_001636057.1)2 from chloroflexus aurantiacus j-10-fl at 1.50 a resolution
|