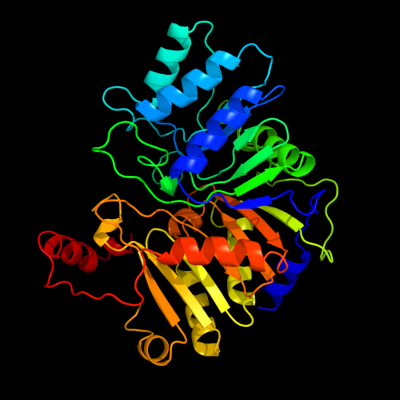
| 1 | c3hsiC_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: C: PDB Molecule:phosphatidylserine synthase;
PDBTitle: crystal structure of phosphatidylserine synthase haemophilus2 influenzae rd kw20
|
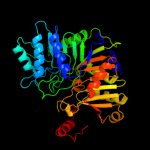
| 2 | c1v0sA_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:phospholipase d;
PDBTitle: uninhibited form of phospholipase d from streptomyces sp.2 strain pmf
|
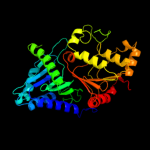
| 3 | c1xdoB_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of escherichia coli polyphosphate kinase
|
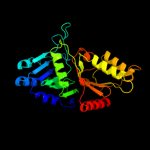
| 4 | d1v0wa1
|
|
 |
100.0 |
15 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Phospholipase D |
| 5 | d1xdpa3
|
|
 |
99.9 |
19 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 6 | d1byra_
|
|
 |
99.9 |
23 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Nuclease |
| 7 | c2o8rA_
|
|
 |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of polyphosphate kinase from2 porphyromonas gingivalis
|
| 8 | d1v0wa2
|
|
 |
99.9 |
23 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Phospholipase D |
| 9 | d2o8ra3
|
|
 |
99.1 |
22 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 10 | d1xdpa4
|
|
 |
99.1 |
15 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 11 | d2o8ra4
|
|
 |
98.9 |
11 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 12 | c1q32C_
|
|
 |
97.7 |
13 |
PDB header:replication,transcription,hydrolase
Chain: C: PDB Molecule:tyrosyl-dna phosphodiesterase;
PDBTitle: crystal structure analysis of the yeast tyrosyl-dna2 phosphodiesterase
|
| 13 | c2c1lA_
|
|
 |
97.6 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:restriction endonuclease;
PDBTitle: structure of the bfii restriction endonuclease
|
| 14 | c3sq3C_
|
|
 |
97.2 |
15 |
PDB header:hydrolase
Chain: C: PDB Molecule:tyrosyl-dna phosphodiesterase 1;
PDBTitle: crystal structure analysis of the yeast tyrosyl-dna phosphodiesterase2 h182a mutant
|
| 15 | d1jy1a2
|
|
 |
95.3 |
18 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 16 | c1nopB_
|
|
 |
95.1 |
23 |
PDB header:hydrolase/dna
Chain: B: PDB Molecule:tyrosyl-dna phosphodiesterase 1;
PDBTitle: crystal structure of human tyrosyl-dna phosphodiesterase2 (tdp1) in complex with vanadate, dna and a human3 topoisomerase i-derived peptide
|
| 17 | d1q32a2
|
|
 |
94.6 |
23 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 18 | d2f5tx2
|
|
 |
81.2 |
23 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:TrmB middle domain-like |
| 19 | c2f5tX_
|
|
 |
75.7 |
23 |
PDB header:transcription
Chain: X: PDB Molecule:archaeal transcriptional regulator trmb;
PDBTitle: crystal structure of the sugar binding domain of the archaeal2 transcriptional regulator trmb
|
| 20 | c3mk7F_
|
|
 |
69.0 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
|
| 21 | d1qzqa1 |
|
not modelled |
66.2 |
36 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 22 | d1o98a1 |
|
not modelled |
65.2 |
10 |
Fold:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
Superfamily:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
Family:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain |
| 23 | d1q32a1 |
|
not modelled |
63.4 |
13 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 24 | d1jy1a1 |
|
not modelled |
61.2 |
8 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
| 25 | c2a5hC_ |
|
not modelled |
56.7 |
18 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
| 26 | c2l82A_ |
|
not modelled |
56.5 |
12 |
PDB header:de novo protein
Chain: A: PDB Molecule:designed protein or32;
PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or32
|
| 27 | d1tfra2 |
|
not modelled |
55.7 |
10 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 28 | c3kgkA_ |
|
not modelled |
54.1 |
11 |
PDB header:chaperone
Chain: A: PDB Molecule:arsenical resistance operon trans-acting repressor arsd;
PDBTitle: crystal structure of arsd
|
| 29 | c2b34C_ |
|
not modelled |
45.3 |
19 |
PDB header:hydrolase
Chain: C: PDB Molecule:mar1 ribonuclease;
PDBTitle: structure of mar1 ribonuclease from caenorhabditis elegans
|
| 30 | c1o98A_ |
|
not modelled |
39.9 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-bisphosphoglycerate-independent
PDBTitle: 1.4a crystal structure of phosphoglycerate mutase from2 bacillus stearothermophilus complexed with3 2-phosphoglycerate
|
| 31 | d1yaca_ |
|
not modelled |
36.5 |
21 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 32 | c3ktbD_ |
|
not modelled |
35.9 |
15 |
PDB header:transcription regulator
Chain: D: PDB Molecule:arsenical resistance operon trans-acting repressor;
PDBTitle: crystal structure of arsenical resistance operon trans-acting2 repressor from bacteroides vulgatus atcc 8482
|
| 33 | c2yv5A_ |
|
not modelled |
35.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:yjeq protein;
PDBTitle: crystal structure of yjeq from aquifex aeolicus
|
| 34 | d2r6gf1 |
|
not modelled |
34.2 |
18 |
Fold:MalF N-terminal region-like
Superfamily:MalF N-terminal region-like
Family:MalF N-terminal region-like |
| 35 | c3gbcA_ |
|
not modelled |
27.9 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrazinamidase/nicotinamidas pnca;
PDBTitle: determination of the crystal structure of the pyrazinamidase from2 m.tuberculosis : a structure-function analysis for prediction3 resistance to pyrazinamide
|
| 36 | d1x9ga_ |
|
not modelled |
27.6 |
14 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 37 | c2nytB_ |
|
not modelled |
27.0 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable c->u-editing enzyme apobec-2;
PDBTitle: the apobec2 crystal structure and functional implications2 for aid
|
| 38 | d1un8a4 |
|
not modelled |
26.9 |
13 |
Fold:DAK1/DegV-like
Superfamily:DAK1/DegV-like
Family:DAK1 |
| 39 | d1gz0a2 |
|
not modelled |
24.7 |
15 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:RNA 2'-O ribose methyltransferase substrate binding domain |
| 40 | c1yzvA_ |
|
not modelled |
23.3 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: hypothetical protein from trypanosoma cruzi
|
| 41 | d1im5a_ |
|
not modelled |
22.7 |
16 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 42 | d1fftb2 |
|
not modelled |
20.5 |
17 |
Fold:Transmembrane helix hairpin
Superfamily:Cytochrome c oxidase subunit II-like, transmembrane region
Family:Cytochrome c oxidase subunit II-like, transmembrane region |
| 43 | c1pt9B_ |
|
not modelled |
20.1 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad(p) transhydrogenase, mitochondrial;
PDBTitle: crystal structure analysis of the diii component of transhydrogenase2 with a thio-nicotinamide nucleotide analogue
|
| 44 | c2r60A_ |
|
not modelled |
20.0 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:glycosyl transferase, group 1;
PDBTitle: structure of apo sucrose phosphate synthase (sps) of2 halothermothrix orenii
|
| 45 | c2bruC_ |
|
not modelled |
20.0 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nad(p) transhydrogenase subunit beta;
PDBTitle: complex of the domain i and domain iii of escherichia coli2 transhydrogenase
|
| 46 | c3l2iB_ |
|
not modelled |
19.8 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
|
| 47 | d1d4oa_ |
|
not modelled |
19.5 |
16 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Transhydrogenase domain III (dIII) |
| 48 | d1pnoa_ |
|
not modelled |
19.4 |
10 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Transhydrogenase domain III (dIII) |
| 49 | d1ueha_ |
|
not modelled |
17.9 |
16 |
Fold:Undecaprenyl diphosphate synthase
Superfamily:Undecaprenyl diphosphate synthase
Family:Undecaprenyl diphosphate synthase |
| 50 | d1gz0f2 |
|
not modelled |
17.8 |
15 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:RNA 2'-O ribose methyltransferase substrate binding domain |
| 51 | c2x7mA_ |
|
not modelled |
17.1 |
5 |
PDB header:hydrolase
Chain: A: PDB Molecule:archaemetzincin;
PDBTitle: crystal structure of archaemetzincin (amza) from methanopyrus2 kandleri at 1.5 a resolution
|
| 52 | c2q5cA_ |
|
not modelled |
16.6 |
9 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
| 53 | c2b8eB_ |
|
not modelled |
16.4 |
13 |
PDB header:membrane protein
Chain: B: PDB Molecule:cation-transporting atpase;
PDBTitle: copa atp binding domain
|
| 54 | c3dy0B_ |
|
not modelled |
16.0 |
40 |
PDB header:blood clotting, hydrolase inhibitor
Chain: B: PDB Molecule:c-terminus plasma serine protease inhibitor;
PDBTitle: crystal structure of cleaved pci bound to heparin
|
| 55 | c2h0rD_ |
|
not modelled |
15.5 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:nicotinamidase;
PDBTitle: structure of the yeast nicotinamidase pnc1p
|
| 56 | c1lq8H_ |
|
not modelled |
14.9 |
40 |
PDB header:blood clotting
Chain: H: PDB Molecule:plasma serine protease inhibitor;
PDBTitle: crystal structure of cleaved protein c inhibitor
|
| 57 | c2pjuD_ |
|
not modelled |
14.9 |
14 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon2 regulatory protein prpr
|
| 58 | c1s21A_ |
|
not modelled |
14.5 |
23 |
PDB header:chaperone
Chain: A: PDB Molecule:orf2;
PDBTitle: crystal structure of avrpphf orf2, a type iii effector from2 p. syringae
|
| 59 | d1s21a_ |
|
not modelled |
14.5 |
23 |
Fold:ADP-ribosylation
Superfamily:ADP-ribosylation
Family:AvrPphF ORF2, a type III effector |
| 60 | c1t3gB_ |
|
not modelled |
13.9 |
11 |
PDB header:membrane protein
Chain: B: PDB Molecule:x-linked interleukin-1 receptor accessory
PDBTitle: crystal structure of the toll/interleukin-1 receptor (tir)2 domain of human il-1rapl
|
| 61 | d1ay7b_ |
|
not modelled |
13.9 |
12 |
Fold:Barstar-like
Superfamily:Barstar-related
Family:Barstar-related |
| 62 | c2wtaA_ |
|
not modelled |
13.8 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:nicotinamidase;
PDBTitle: acinetobacter baumanii nicotinamidase pyrazinamidease
|
| 63 | c2yvkA_ |
|
not modelled |
13.8 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
|
| 64 | c3hu5B_ |
|
not modelled |
13.7 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:isochorismatase family protein;
PDBTitle: crystal structure of isochorismatase family protein from desulfovibrio2 vulgaris subsp. vulgaris str. hildenborough
|
| 65 | d1x94a_ |
|
not modelled |
13.6 |
15 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
| 66 | d1pjqa2 |
|
not modelled |
13.6 |
13 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 67 | d1o57a2 |
|
not modelled |
13.5 |
14 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 68 | d1t5oa_ |
|
not modelled |
13.4 |
13 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 69 | c3js3C_ |
|
not modelled |
13.4 |
8 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of type i 3-dehydroquinate dehydratase (arod) from2 clostridium difficile with covalent reaction intermediate
|
| 70 | c2rddB_ |
|
not modelled |
13.4 |
17 |
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
|
| 71 | d1nvmb1 |
|
not modelled |
13.4 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 72 | d3dtub2 |
|
not modelled |
13.3 |
7 |
Fold:Transmembrane helix hairpin
Superfamily:Cytochrome c oxidase subunit II-like, transmembrane region
Family:Cytochrome c oxidase subunit II-like, transmembrane region |
| 73 | c3igzB_ |
|
not modelled |
13.2 |
9 |
PDB header:isomerase
Chain: B: PDB Molecule:cofactor-independent phosphoglycerate mutase;
PDBTitle: crystal structures of leishmania mexicana phosphoglycerate2 mutase at low cobalt concentration
|
| 74 | c2iksA_ |
|
not modelled |
13.0 |
8 |
PDB header:transcription
Chain: A: PDB Molecule:dna-binding transcriptional dual regulator;
PDBTitle: crystal structure of n-terminal truncated dna-binding transcriptional2 dual regulator from escherichia coli k12
|
| 75 | d2fcja1 |
|
not modelled |
12.9 |
7 |
Fold:Toprim domain
Superfamily:Toprim domain
Family:Toprim domain |
| 76 | d1t9ka_ |
|
not modelled |
12.7 |
20 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 77 | d1s4da_ |
|
not modelled |
12.6 |
10 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 78 | c3r2jC_ |
|
not modelled |
12.5 |
15 |
PDB header:hydrolase
Chain: C: PDB Molecule:alpha/beta-hydrolase-like protein;
PDBTitle: crystal structure of pnc1 from l. infantum in complex with nicotinate
|
| 79 | c2hqbA_ |
|
not modelled |
12.2 |
8 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional activator of comk gene;
PDBTitle: crystal structure of a transcriptional activator of comk2 gene from bacillus halodurans
|
| 80 | c2voyC_ |
|
not modelled |
12.2 |
38 |
PDB header:hydrolase
Chain: C: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
|
| 81 | c3brqA_ |
|
not modelled |
12.0 |
5 |
PDB header:transcription regulator
Chain: A: PDB Molecule:hth-type transcriptional regulator ascg;
PDBTitle: crystal structure of the escherichia coli transcriptional repressor2 ascg
|
| 82 | c3zvmA_ |
|
not modelled |
11.9 |
20 |
PDB header:hydrolase/transferase/dna
Chain: A: PDB Molecule:bifunctional polynucleotide phosphatase/kinase;
PDBTitle: the structural basis for substrate recognition by mammalian2 polynucleotide kinase 3' phosphatase
|
| 83 | c1hf2A_ |
|
not modelled |
11.8 |
14 |
PDB header:cell division protein
Chain: A: PDB Molecule:septum site-determining protein minc;
PDBTitle: crystal structure of the bacterial cell-division inhibitor2 minc from t. maritima
|
| 84 | c3a11D_ |
|
not modelled |
11.7 |
10 |
PDB header:isomerase
Chain: D: PDB Molecule:translation initiation factor eif-2b, delta
PDBTitle: crystal structure of ribose-1,5-bisphosphate isomerase from2 thermococcus kodakaraensis kod1
|
| 85 | c3h75A_ |
|
not modelled |
11.6 |
7 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:periplasmic sugar-binding domain protein;
PDBTitle: crystal structure of a periplasmic sugar-binding protein from the2 pseudomonas fluorescens
|
| 86 | d1acoa2 |
|
not modelled |
11.5 |
10 |
Fold:Aconitase iron-sulfur domain
Superfamily:Aconitase iron-sulfur domain
Family:Aconitase iron-sulfur domain |
| 87 | d1gpja3 |
|
not modelled |
11.4 |
15 |
Fold:Ferredoxin-like
Superfamily:Glutamyl tRNA-reductase catalytic, N-terminal domain
Family:Glutamyl tRNA-reductase catalytic, N-terminal domain |
| 88 | d1nr3a_ |
|
not modelled |
11.4 |
21 |
Fold:DNA-binding protein Tfx
Superfamily:DNA-binding protein Tfx
Family:DNA-binding protein Tfx |
| 89 | c2rchA_ |
|
not modelled |
11.3 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:cytochrome p450 74a;
PDBTitle: crystal structure of arabidopsis thaliana allene oxide synthase (aos,2 cytochrome p450 74a, cyp74a) complexed with 13(s)-hod at 1.85 a3 resolution
|
| 90 | d1ecfa1 |
|
not modelled |
11.0 |
17 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 91 | c3ny7A_ |
|
not modelled |
10.9 |
13 |
PDB header:membrane protein
Chain: A: PDB Molecule:sulfate transporter;
PDBTitle: stas domain of ychm bound to acp
|
| 92 | d1j5pa4 |
|
not modelled |
10.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 93 | c3h5oB_ |
|
not modelled |
10.7 |
15 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator gntr;
PDBTitle: the crystal structure of transcription regulator gntr from2 chromobacterium violaceum
|
| 94 | c3ecsD_ |
|
not modelled |
10.5 |
13 |
PDB header:translation
Chain: D: PDB Molecule:translation initiation factor eif-2b subunit
PDBTitle: crystal structure of human eif2b alpha
|
| 95 | c3bblA_ |
|
not modelled |
10.4 |
11 |
PDB header:regulatory protein
Chain: A: PDB Molecule:regulatory protein of laci family;
PDBTitle: crystal structure of a regulatory protein of laci family from2 chloroflexus aggregans
|
| 96 | d1qyda_ |
|
not modelled |
10.2 |
6 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 97 | d1gqna_ |
|
not modelled |
10.0 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 98 | c1o57A_ |
|
not modelled |
10.0 |
14 |
PDB header:dna binding protein
Chain: A: PDB Molecule:pur operon repressor;
PDBTitle: crystal structure of the purine operon repressor of2 bacillus subtilis
|
| 99 | d1wdea_ |
|
not modelled |
10.0 |
14 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |