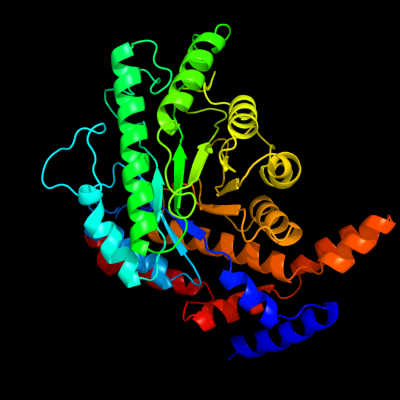
| 1 | d1d8wa_
|
|
 |
100.0 |
98 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:L-rhamnose isomerase |
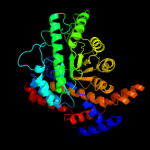
| 2 | c3p14C_
|
|
 |
100.0 |
59 |
PDB header:isomerase
Chain: C: PDB Molecule:l-rhamnose isomerase;
PDBTitle: crystal structure of l-rhamnose isomerase with a novel high thermo-2 stability from bacillus halodurans
|
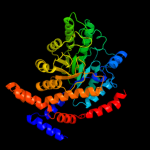
| 3 | c2i56A_
|
|
 |
100.0 |
19 |
PDB header:isomerase, metal-binding protein
Chain: A: PDB Molecule:l-rhamnose isomerase;
PDBTitle: crystal structure of l-rhamnose isomerase from pseudomonas2 stutzeri with l-rhamnose
|
| 4 | d1xlma_
|
|
 |
100.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 5 | d1bxca_
|
|
 |
100.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 6 | d1bxba_
|
|
 |
100.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 7 | c3ktcB_
|
|
 |
100.0 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_050048.1) from2 erwinia carotovora atroseptica scri1043 at 1.54 a resolution
|
| 8 | d2glka1
|
|
 |
100.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 9 | d1muwa_
|
|
 |
100.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 10 | d1qt1a_
|
|
 |
100.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 11 | d1xima_
|
|
 |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 12 | d1a0ea_
|
|
 |
99.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 13 | d1a0da_
|
|
 |
99.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 14 | d1a0ca_
|
|
 |
99.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 15 | c3qxbB_
|
|
 |
98.7 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:putative xylose isomerase;
PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
|
| 16 | c2qw5B_
|
|
 |
98.6 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase-like tim barrel;
PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
|
| 17 | c2zvrA_
|
|
 |
98.5 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:uncharacterized protein tm_0416;
PDBTitle: crystal structure of a d-tagatose 3-epimerase-related2 protein from thermotoga maritima
|
| 18 | c3cqkB_
|
|
 |
98.5 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose-5-phosphate 3-epimerase ulae;
PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
|
| 19 | c3kwsB_
|
|
 |
98.2 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
|
| 20 | c2hk1D_
|
|
 |
98.2 |
13 |
PDB header:isomerase
Chain: D: PDB Molecule:d-psicose 3-epimerase;
PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
|
| 21 | d1i60a_ |
|
not modelled |
98.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 22 | d1xp3a1 |
|
not modelled |
98.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Endonuclease IV |
| 23 | c2x7vA_ |
|
not modelled |
98.1 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable endonuclease 4;
PDBTitle: crystal structure of thermotoga maritima endonuclease iv in2 the presence of zinc
|
| 24 | d1qtwa_ |
|
not modelled |
98.0 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Endonuclease IV |
| 25 | c3ju2A_ |
|
not modelled |
97.9 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein smc04130;
PDBTitle: crystal structure of protein smc04130 from sinorhizobium meliloti 1021
|
| 26 | d1k77a_ |
|
not modelled |
97.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Hypothetical protein YgbM (EC1530) |
| 27 | c3bzjA_ |
|
not modelled |
97.7 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:uv endonuclease;
PDBTitle: uvde k229l
|
| 28 | c2zdsB_ |
|
not modelled |
97.7 |
11 |
PDB header:dna binding protein
Chain: B: PDB Molecule:putative dna-binding protein;
PDBTitle: crystal structure of sco6571 from streptomyces coelicolor2 a3(2)
|
| 29 | c3obeB_ |
|
not modelled |
97.7 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_3400)2 from parabacteroides distasonis atcc 8503 at 1.70 a resolution
|
| 30 | c2ou4C_ |
|
not modelled |
97.5 |
14 |
PDB header:isomerase
Chain: C: PDB Molecule:d-tagatose 3-epimerase;
PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
|
| 31 | c3aamA_ |
|
not modelled |
97.3 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease iv;
PDBTitle: crystal structure of endonuclease iv from thermus thermophilus hb8
|
| 32 | c3ngfA_ |
|
not modelled |
97.1 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:ap endonuclease, family 2;
PDBTitle: crystal structure of ap endonuclease, family 2 from brucella2 melitensis
|
| 33 | c3dx5A_ |
|
not modelled |
97.1 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
| 34 | c3l23A_ |
|
not modelled |
97.0 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of sugar phosphate isomerase/epimerase2 (yp_001303399.1) from parabacteroides distasonis atcc 8503 at 1.70 a3 resolution
|
| 35 | c3lmzA_ |
|
not modelled |
96.7 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase. (yp_001305105.1) from2 parabacteroides distasonis atcc 8503 at 1.44 a resolution
|
| 36 | d1tz9a_ |
|
not modelled |
96.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:UxuA-like |
| 37 | c3cnyA_ |
|
not modelled |
96.5 |
12 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:inositol catabolism protein iole;
PDBTitle: crystal structure of a putative inositol catabolism protein iole2 (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 a3 resolution
|
| 38 | c3bdkB_ |
|
not modelled |
96.4 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:d-mannonate dehydratase;
PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
|
| 39 | d2g0wa1 |
|
not modelled |
96.0 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 40 | d2q02a1 |
|
not modelled |
93.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
| 41 | d1yx1a1 |
|
not modelled |
93.3 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:KguE-like |
| 42 | c3p6lA_ |
|
not modelled |
89.6 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_1903)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
|
| 43 | c2wvsD_ |
|
not modelled |
84.8 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:alpha-l-fucosidase;
PDBTitle: crystal structure of an alpha-l-fucosidase gh29 trapped2 covalent intermediate from bacteroides thetaiotaomicron in3 complex with 2-fluoro-fucosyl fluoride using an e288q4 mutant
|
| 44 | d1x7fa2 |
|
not modelled |
73.7 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Outer surface protein, N-terminal domain |
| 45 | c2p0oA_ |
|
not modelled |
70.6 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein duf871;
PDBTitle: crystal structure of a conserved protein from locus ef_2437 in2 enterococcus faecalis with an unknown function
|
| 46 | c1uasA_ |
|
not modelled |
64.7 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase;
PDBTitle: crystal structure of rice alpha-galactosidase
|
| 47 | c2vpnB_ |
|
not modelled |
55.6 |
14 |
PDB header:transport
Chain: B: PDB Molecule:periplasmic substrate binding protein;
PDBTitle: high-resolution structure of the periplasmic ectoine-2 binding protein from teaabc trap-transporter of halomonas3 elongata
|
| 48 | c2by0A_ |
|
not modelled |
54.6 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:maltooligosyltrehalose trehalohydrolase;
PDBTitle: is radiation damage dependent on the dose-rate used during2 macromolecular crystallography data collection
|
| 49 | c3pt1A_ |
|
not modelled |
50.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:upf0364 protein ymr027w;
PDBTitle: structure of duf89 from saccharomyces cerevisiae co-crystallized with2 f6p.
|
| 50 | d1ktba2 |
|
not modelled |
48.2 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 51 | c1x7fA_ |
|
not modelled |
48.0 |
32 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:outer surface protein;
PDBTitle: crystal structure of an uncharacterized b. cereus protein
|
| 52 | c2d0gA_ |
|
not modelled |
45.5 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-amylase i;
PDBTitle: crystal structure of thermoactinomyces vulgaris r-47 alpha-2 amylase 1 (tvai) mutant d356n/e396q complexed with p5, a3 pullulan model oligosaccharide
|
| 53 | d1uasa2 |
|
not modelled |
42.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 54 | c3lrmB_ |
|
not modelled |
41.3 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-galactosidase 1;
PDBTitle: structure of alfa-galactosidase from saccharomyces cerevisiae with2 raffinose
|
| 55 | c2ya0A_ |
|
not modelled |
38.2 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative alkaline amylopullulanase;
PDBTitle: catalytic module of the multi-modular glycogen-degrading2 pneumococcal virulence factor spua
|
| 56 | c3emzA_ |
|
not modelled |
37.6 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-1,4-beta-xylanase;
PDBTitle: crystal structure of xylanase xynb from paenibacillus2 barcinonensis complexed with a conduramine derivative
|
| 57 | d1kwga2 |
|
not modelled |
37.6 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 58 | d1q2ha_ |
|
not modelled |
36.9 |
15 |
Fold:Dimerisation interlock
Superfamily:Phenylalanine zipper
Family:Adapter protein APS, dimerisation domain |
| 59 | c3czkA_ |
|
not modelled |
36.4 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:sucrose hydrolase;
PDBTitle: crystal structure analysis of sucrose hydrolase(suh) e322q-2 sucrose complex
|
| 60 | c3k1dA_ |
|
not modelled |
34.3 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:1,4-alpha-glucan-branching enzyme;
PDBTitle: crystal structure of glycogen branching enzyme synonym: 1,4-alpha-d-2 glucan:1,4-alpha-d-glucan 6-glucosyl-transferase from mycobacterium3 tuberculosis h37rv
|
| 61 | d2fywa1 |
|
not modelled |
33.7 |
26 |
Fold:NIF3 (NGG1p interacting factor 3)-like
Superfamily:NIF3 (NGG1p interacting factor 3)-like
Family:NIF3 (NGG1p interacting factor 3)-like |
| 62 | d1bf2a3 |
|
not modelled |
33.6 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 63 | c3r66A_ |
|
not modelled |
33.2 |
20 |
PDB header:viral protein/antiviral protein
Chain: A: PDB Molecule:non-structural protein 1;
PDBTitle: crystal structure of human isg15 in complex with ns1 n-terminal region2 from influenza virus b, northeast structural genomics consortium3 target ids hx6481, hr2873, and or2
|
| 64 | d1cyga4 |
|
not modelled |
33.1 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 65 | d1xeqa1 |
|
not modelled |
33.0 |
20 |
Fold:S15/NS1 RNA-binding domain
Superfamily:S15/NS1 RNA-binding domain
Family:N-terminal, RNA-binding domain of nonstructural protein NS1 |
| 66 | c1h2aS_ |
|
not modelled |
32.5 |
24 |
PDB header:oxidoreductase
Chain: S: PDB Molecule:hydrogenase;
PDBTitle: single crystals of hydrogenase from desulfovibrio vulgaris
|
| 67 | c2if8B_ |
|
not modelled |
32.2 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:inositol polyphosphate multikinase;
PDBTitle: crystal structure of inositol phosphate multikinase ipk2 in complex2 with adp and mn2+ from s. cerevisiae
|
| 68 | c3m07A_ |
|
not modelled |
31.9 |
26 |
PDB header:unknown function
Chain: A: PDB Molecule:putative alpha amylase;
PDBTitle: 1.4 angstrom resolution crystal structure of putative alpha2 amylase from salmonella typhimurium.
|
| 69 | c3a23A_ |
|
not modelled |
31.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative secreted alpha-galactosidase;
PDBTitle: crystal structure of beta-l-arabinopyranosidase complexed with d-2 galactose
|
| 70 | d1jvaa2 |
|
not modelled |
31.0 |
32 |
Fold:Homing endonuclease-like
Superfamily:Homing endonucleases
Family:Intein endonuclease |
| 71 | d1wuis1 |
|
not modelled |
30.7 |
24 |
Fold:HydA/Nqo6-like
Superfamily:HydA/Nqo6-like
Family:Nickel-iron hydrogenase, small subunit |
| 72 | c1ktbA_ |
|
not modelled |
30.7 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-n-acetylgalactosaminidase;
PDBTitle: the structure of alpha-n-acetylgalactosaminidase
|
| 73 | d1wzaa2 |
|
not modelled |
30.5 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 74 | c3rgwS_ |
|
not modelled |
30.3 |
23 |
PDB header:oxidoreductase/oxidoreductase
Chain: S: PDB Molecule:membrane-bound hydrogenase (nife) small subunit hoxk;
PDBTitle: crystal structure at 1.5 a resolution of an h2-reduced, o2-tolerant2 hydrogenase from ralstonia eutropha unmasks a novel iron-sulfur3 cluster
|
| 75 | c2vncB_ |
|
not modelled |
29.9 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycogen operon protein glgx;
PDBTitle: crystal structure of glycogen debranching enzyme trex from2 sulfolobus solfataricus
|
| 76 | d1e3da_ |
|
not modelled |
29.9 |
32 |
Fold:HydA/Nqo6-like
Superfamily:HydA/Nqo6-like
Family:Nickel-iron hydrogenase, small subunit |
| 77 | d1y0na_ |
|
not modelled |
28.3 |
19 |
Fold:YehU-like
Superfamily:YehU-like
Family:YehU-like |
| 78 | d2aaaa2 |
|
not modelled |
27.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 79 | d1uoza_ |
|
not modelled |
27.5 |
17 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycosyl hydrolases family 6, cellulases
Family:Glycosyl hydrolases family 6, cellulases |
| 80 | c3myrE_ |
|
not modelled |
26.9 |
25 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:hydrogenase (nife) small subunit hyda;
PDBTitle: crystal structure of [nife] hydrogenase from allochromatium vinosum in2 its ni-a state
|
| 81 | c3a5vA_ |
|
not modelled |
26.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase;
PDBTitle: crystal structure of alpha-galactosidase i from mortierella vinacea
|
| 82 | d1eh9a3 |
|
not modelled |
26.3 |
31 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 83 | d1r46a2 |
|
not modelled |
26.3 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 84 | c2kelB_ |
|
not modelled |
25.6 |
17 |
PDB header:transcription repressor
Chain: B: PDB Molecule:uncharacterized protein 56b;
PDBTitle: structure of the transcription regulator svtr from the2 hyperthermophilic archaeal virus sirv1
|
| 85 | c2e8yA_ |
|
not modelled |
25.2 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:amyx protein;
PDBTitle: crystal structure of pullulanase type i from bacillus subtilis str.2 168
|
| 86 | c1wzaA_ |
|
not modelled |
25.1 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-amylase a;
PDBTitle: crystal structure of alpha-amylase from h.orenii
|
| 87 | d1tjoa_ |
|
not modelled |
25.1 |
11 |
Fold:Ferritin-like
Superfamily:Ferritin-like
Family:Ferritin |
| 88 | d1frfs_ |
|
not modelled |
24.9 |
32 |
Fold:HydA/Nqo6-like
Superfamily:HydA/Nqo6-like
Family:Nickel-iron hydrogenase, small subunit |
| 89 | c3uowB_ |
|
not modelled |
24.8 |
7 |
PDB header:ligase
Chain: B: PDB Molecule:gmp synthetase;
PDBTitle: crystal structure of pf10_0123, a gmp synthetase from plasmodium2 falciparum
|
| 90 | d1yq9a1 |
|
not modelled |
24.4 |
24 |
Fold:HydA/Nqo6-like
Superfamily:HydA/Nqo6-like
Family:Nickel-iron hydrogenase, small subunit |
| 91 | c3qomA_ |
|
not modelled |
24.2 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phospho-beta-glucosidase;
PDBTitle: crystal structure of 6-phospho-beta-glucosidase from lactobacillus2 plantarum
|
| 92 | c2nydB_ |
|
not modelled |
23.7 |
13 |
PDB header:unknown function
Chain: B: PDB Molecule:upf0135 protein sa1388;
PDBTitle: crystal structure of staphylococcus aureus hypothetical protein sa1388
|
| 93 | c2kebA_ |
|
not modelled |
23.1 |
24 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna polymerase subunit alpha b;
PDBTitle: nmr solution structure of the n-terminal domain of the dna polymerase2 alpha p68 subunit
|
| 94 | c1t0oA_ |
|
not modelled |
22.4 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase;
PDBTitle: the structure of alpha-galactosidase from trichoderma reesei complexed2 with beta-d-galactose
|
| 95 | d1z8ma1 |
|
not modelled |
22.2 |
12 |
Fold:RelE-like
Superfamily:RelE-like
Family:RelE-like |
| 96 | d1xyna_ |
|
not modelled |
22.0 |
33 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Xylanase/endoglucanase 11/12 |
| 97 | c3civA_ |
|
not modelled |
21.3 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-beta-1,4-mannanase;
PDBTitle: crystal structure of the endo-beta-1,4-mannanase from2 alicyclobacillus acidocaldarius
|
| 98 | c3uj2C_ |
|
not modelled |
20.9 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:enolase 1;
PDBTitle: crystal structure of an enolase from anaerostipes caccae (efi target2 efi-502054) with bound mg and sulfate
|
| 99 | c2cncA_ |
|
not modelled |
20.8 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoxylanase;
PDBTitle: family 10 xylanase
|
| 100 | c3eypB_ |
|
not modelled |
20.8 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of putative alpha-l-fucosidase from bacteroides2 thetaiotaomicron
|
| 101 | c3d1nK_ |
|
not modelled |
20.2 |
26 |
PDB header:transcription regulator/dna
Chain: K: PDB Molecule:pou domain, class 6, transcription factor 1;
PDBTitle: structure of human brn-5 transcription factor in complex2 with corticotrophin-releasing hormone gene promoter
|
| 102 | d1xyza_ |
|
not modelled |
20.0 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 103 | c1xyzA_ |
|
not modelled |
20.0 |
11 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:1,4-beta-d-xylan-xylanohydrolase;
PDBTitle: a common protein fold and similar active site in two2 distinct families of beta-glycanases
|