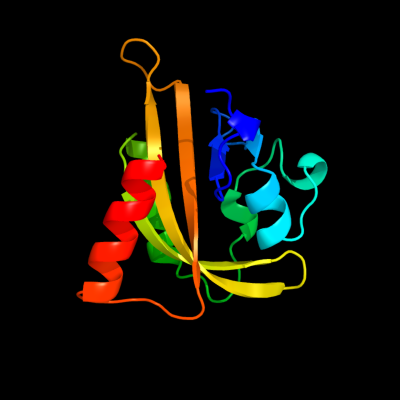
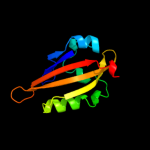
1 c2wkqA_
99.9
20
PDB header: transferase, cell adhesionChain: A: PDB Molecule: nph1-1, ras-related c3 botulinum toxin substratePDBTitle: structure of a photoactivatable rac1 containing the lov22 c450a mutant
2 d1p97a_
99.8
19
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Hypoxia-inducible factor Hif2a, C-terminal domain3 c3rtyA_
99.8
6
PDB header: circadian clock proteinChain: A: PDB Molecule: period circadian protein;PDBTitle: structure of an enclosed dimer formed by the drosophila period protein
4 d1jnua_
99.8
19
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Flavin-binding PAS domain5 c2z6dB_
99.8
20
PDB header: transferaseChain: B: PDB Molecule: phototropin-2;PDBTitle: crystal structure of lov1 domain of phototropin2 from2 arabidopsis thaliana
6 d1bywa_
99.7
18
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Flavin-binding PAS domain7 c3f1oB_
99.7
21
PDB header: transcriptionChain: B: PDB Molecule: aryl hydrocarbon receptor nuclear translocator;PDBTitle: crystal structure of the high affinity heterodimer of hif22 alpha and arnt c-terminal pas domains, with an internally-3 bound artificial ligand
8 c2v1bA_
99.7
20
PDB header: transferaseChain: A: PDB Molecule: nph1-1;PDBTitle: n- and c-terminal helices of oat lov2 (404-546) are2 involved in light-induced signal transduction (room3 temperature (293k) light structure of lov2 (404-546))
9 c2kdkA_
99.7
15
PDB header: transcription regulatorChain: A: PDB Molecule: aryl hydrocarbon receptor nuclear translocator-like proteinPDBTitle: structure of human circadian clock protein bmal2 c-terminal pas domain
10 d1n9la_
99.7
22
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Flavin-binding PAS domain11 c3ewkA_
99.7
30
PDB header: flavoproteinChain: A: PDB Molecule: sensor protein;PDBTitle: structure of the redox sensor domain of methylococcus capsulatus2 (bath) mmos
12 c2vlgD_
99.7
13
PDB header: transferaseChain: D: PDB Molecule: sporulation kinase a;PDBTitle: kina pas-a domain, homodimer
13 c3gdiB_
99.7
17
PDB header: transcriptionChain: B: PDB Molecule: period circadian protein homolog 2;PDBTitle: mammalian clock protein mper2 - crystal struture of a pas2 domain fragment
14 c2pr6A_
99.7
22
PDB header: flavoprotein, signaling proteinChain: A: PDB Molecule: blue-light photoreceptor;PDBTitle: structural basis for light-dependent signaling in the dimeric lov2 photosensor ytva (light structure)
15 c2l4rA_
99.7
15
PDB header: transport proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily h member 2;PDBTitle: nmr solution structure of the n-terminal pas domain of herg
16 d1oj5a_
99.7
12
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: PAS domain of steroid receptor coactivator 1A, NCo-A117 c3mjqB_
99.7
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the pas domain of q24qt8_deshy protein from2 desulfitobacterium hafniense. northeast structural genomics3 consortium target dhr85c.
18 c3lyxA_
99.6
22
PDB header: transcriptionChain: A: PDB Molecule: sensory box/ggdef domain protein;PDBTitle: crystal structure of the pas domain of the protein cps_12912 from colwellia psychrerythraea. northeast structural3 genomics consortium target id csr222b
19 c2gj3A_
99.6
24
PDB header: transferaseChain: A: PDB Molecule: nitrogen fixation regulatory protein;PDBTitle: crystal structure of the fad-containing pas domain of the2 protein nifl from azotobacter vinelandii.
20 c3h9wA_
99.6
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: diguanylate cyclase with pas/pac sensor;PDBTitle: crystal structure of the n-terminal domain of diguanylate cyclase with2 pas/pac sensor (maqu_2914) from marinobacter aquaeolei, northeast3 structural genomics consortium target mqr66c
21 c3oloB_
not modelled
99.6
10
PDB header: transferaseChain: B: PDB Molecule: two-component sensor histidine kinase;PDBTitle: crystal structure of a pas domain from two-component sensor histidine2 kinase
22 c2pdtD_
not modelled
99.6
19
PDB header: circadian clock proteinChain: D: PDB Molecule: vivid pas protein vvd;PDBTitle: 2.3 angstrom structure of phosphodiesterase treated vivid
23 c3mqoB_
not modelled
99.6
11
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, luxr family;PDBTitle: the crystal structure of the pas domain in complex with isopropanol of2 a transcriptional regulator in the luxr family from burkholderia3 thailandensis to 1.7a
24 c3b33A_
not modelled
99.6
12
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: crystal structure of the pas domain of nitrogen regulation protein2 nr(ii) from vibrio parahaemolyticus
25 d1xj3a1
not modelled
99.6
16
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Heme-binding PAS domain26 c1v9yA_
not modelled
99.6
17
PDB header: signaling proteinChain: A: PDB Molecule: heme pas sensor protein;PDBTitle: crystal structure of the heme pas sensor domain of ec dos (ferric2 form)
27 d1v9ya_
not modelled
99.6
17
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Heme-binding PAS domain28 c3caxA_
not modelled
99.6
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pf0695;PDBTitle: crystal structure of uncharacterized protein pf0695
29 c3gecA_
not modelled
99.6
11
PDB header: circadian clock proteinChain: A: PDB Molecule: period circadian protein;PDBTitle: crystal structure of a tandem pas domain fragment of2 drosophila period
30 c1wa9A_
not modelled
99.6
9
PDB header: circadian rhythmChain: A: PDB Molecule: period circadian protein;PDBTitle: crystal structure of the pas repeat region of the2 drosophila clock protein period
31 c3mfxA_
not modelled
99.6
10
PDB header: transcriptionChain: A: PDB Molecule: sensory box/ggdef family protein;PDBTitle: crystal structure of the sensory box domain of the sensory-2 box/ggdef protein so_1695 from shewanella oneidensis,3 northeast structural genomics consortium target sor288b
32 d1ew0a_
not modelled
99.5
17
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Heme-binding PAS domain33 d1y28a_
not modelled
99.5
14
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Heme-binding PAS domain34 c3fg8B_
not modelled
99.5
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein rha05790;PDBTitle: crystal structure of pas domain of rha05790
35 c3luqC_
not modelled
99.5
14
PDB header: transferaseChain: C: PDB Molecule: sensor protein;PDBTitle: the crystal structure of a pas domain from a sensory box2 histidine kinase regulator from geobacter sulfurreducens to3 2.5a
36 c3p7nB_
not modelled
99.5
11
PDB header: dna binding proteinChain: B: PDB Molecule: sensor histidine kinase;PDBTitle: crystal structure of light activated transcription factor el222 from2 erythrobacter litoralis
37 c2r78D_
not modelled
99.5
17
PDB header: transferaseChain: D: PDB Molecule: sensor protein;PDBTitle: crystal structure of a domain of the sensory box sensor2 histidine kinase/response regulator from geobacter3 sulfurreducens
38 c3bwlA_
not modelled
99.5
14
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: crystal structure of pas domain of htr-like protein from haloarcula2 marismortui
39 c3mr0B_
not modelled
99.5
14
PDB header: transcription regulatorChain: B: PDB Molecule: sensory box histidine kinase/response regulator;PDBTitle: crystal structure of sensory box histidine kinase/response regulator2 from burkholderia thailandensis e264
40 c3fc7B_
not modelled
99.5
12
PDB header: transferaseChain: B: PDB Molecule: htr-like protein;PDBTitle: the crystal structure of a domain of htr-like protein from haloarcula2 marismortui atcc 43049
41 d1ll8a_
not modelled
99.4
18
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: N-terminal PAS domain of Pas kinase42 c3a0vA_
not modelled
99.4
13
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: pas domain of histidine kinase thka (tm1359) (semet,2 f486m/f489m)
43 c3eehA_
not modelled
99.4
14
PDB header: transferaseChain: A: PDB Molecule: putative light and redox sensing histidine kinase;PDBTitle: the crystal structure of the domain of the putative light and redox2 sensing histidine kinase from haloarcula marismortui
44 c1qu7A_
99.4
51
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein i;PDBTitle: four helical-bundle structure of the cytoplasmic domain of a serine2 chemotaxis receptor
45 c3njaC_
not modelled
99.4
10
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: probable ggdef family protein;PDBTitle: the crystal structure of the pas domain of a ggdef family protein from2 chromobacterium violaceum atcc 12472.
46 d1nwza_
not modelled
99.3
9
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: PYP-like47 d1otda_
not modelled
99.3
10
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: PYP-like48 c3mxqC_
not modelled
99.3
12
PDB header: transferaseChain: C: PDB Molecule: sensor protein;PDBTitle: crystal structure of sensory box sensor histidine kinase from vibrio2 cholerae
49 d1mzua_
not modelled
99.3
11
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: PYP-like50 c3icyB_
not modelled
99.3
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: sensor protein;PDBTitle: the crystal structure of sensory box histidine2 kinase/response regulator domain from chlorobium tepidum3 tls
51 d1xfna1
not modelled
99.2
9
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: PYP-like52 c2oolA_
not modelled
99.2
8
PDB header: signaling proteinChain: A: PDB Molecule: sensor protein;PDBTitle: crystal structure of the chromophore-binding domain of an unusual2 bacteriophytochrome rpbphp3 from r. palustris
53 c3k3dA_
not modelled
99.2
18
PDB header: signaling proteinChain: A: PDB Molecule: protein rv1364c/mt1410;PDBTitle: the n-terminal pas domain crystal structure of rv1364c from2 mycobacterium tuberculosis at 2.3 angstrom
54 c2o9bA_
not modelled
99.2
12
PDB header: transferaseChain: A: PDB Molecule: bacteriophytochrome;PDBTitle: crystal structure of bacteriophytochrome chromophore binding domain
55 c3a0rA_
not modelled
99.2
11
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
56 c2qkpD_
not modelled
99.1
11
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of c-terminal domain of smu_1151c from streptococcus2 mutans
57 c2jheB_
not modelled
99.0
13
PDB header: transcriptionChain: B: PDB Molecule: transcription regulator tyrr;PDBTitle: n-terminal domain of tyrr transcription factor (residues 1 -2 190)
58 c3kx0X_
not modelled
99.0
19
PDB header: signaling proteinChain: X: PDB Molecule: uncharacterized protein rv1364c/mt1410;PDBTitle: crystal structure of the pas domain of rv1364c
59 c2w0nA_
not modelled
98.9
19
PDB header: transferaseChain: A: PDB Molecule: sensor protein dcus;PDBTitle: plasticity of pas domain and potential role for signal2 transduction in the histidine-kinase dcus
60 c2veaA_
not modelled
98.9
8
PDB header: transferaseChain: A: PDB Molecule: phytochrome-like protein cph1;PDBTitle: the complete sensory module of the cyanobacterial2 phytochrome cph1 in the pr-state.
61 c2ch7A_
98.9
22
PDB header: chemotaxisChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of the cytoplasmic domain of a bacterial2 chemoreceptor from thermotoga maritima
62 c3cloC_
not modelled
98.7
10
PDB header: transcription regulatorChain: C: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator containing a2 luxr dna binding domain (np_811094.1) from bacteroides3 thetaiotaomicron vpi-5482 at 2.04 a resolution
63 c3g67A_
not modelled
98.3
20
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of a soluble chemoreceptor from thermotoga2 maritima
64 c3c2wB_
not modelled
97.8
14
PDB header: signaling proteinChain: B: PDB Molecule: bacteriophytochrome;PDBTitle: crystal structure of the photosensory core domain of p.2 aeruginosa bacteriophytochrome pabphp in the pfr state
65 c3lnrA_
97.4
8
PDB header: signaling proteinChain: A: PDB Molecule: aerotaxis transducer aer2;PDBTitle: crystal structure of poly-hamp domains from the p. aeruginosa soluble2 receptor aer2
66 d2oola2
not modelled
97.2
7
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: BphP N-terminal domain-like67 d2o9ca2
not modelled
96.6
13
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: BphP N-terminal domain-like68 d3c2wa3
not modelled
95.9
16
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: BphP N-terminal domain-like69 d2veaa3
not modelled
95.6
12
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: BphP N-terminal domain-like70 d2asxa1
95.2
29
Fold: HAMP domain-likeSuperfamily: HAMP domain-likeFamily: HAMP domain71 c2rm8A_
not modelled
93.8
19
PDB header: signaling proteinChain: A: PDB Molecule: sensory rhodopsin ii transducer;PDBTitle: the solution structure of phototactic transducer protein2 htrii linker region from natronomonas pharaonis
72 c3zrwB_
not modelled
92.2
19
PDB header: signaling proteinChain: B: PDB Molecule: af1503 protein, osmolarity sensor protein envz;PDBTitle: the structure of the dimeric hamp-dhp fusion a291v mutant
73 c2wpqA_
not modelled
89.2
6
PDB header: membrane proteinChain: A: PDB Molecule: trimeric autotransporter adhesin fragment;PDBTitle: salmonella enterica sada 479-519 fused to gcn4 adaptors (2 sadak3, in-register fusion)
74 d1p0za_
not modelled
88.9
19
Fold: Profilin-likeSuperfamily: Sensory domain-likeFamily: Sensory domain of two-component sensor kinase75 c1ei3E_
not modelled
88.7
9
PDB header: PDB COMPND: 76 c3ghgK_
not modelled
78.7
14
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
77 c1deqO_
not modelled
78.1
15
PDB header: PDB COMPND: 78 c3fosA_
not modelled
77.7
11
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: crystal structure of two-component sensor histidine kinase domain from2 bacillus subtilis subsp. subtilis str. 168
79 c1sfcJ_
not modelled
75.1
3
PDB header: transport proteinChain: J: PDB Molecule: protein (syntaxin 1a);PDBTitle: neuronal synaptic fusion complex
80 c1ei3C_
not modelled
74.7
7
PDB header: PDB COMPND: 81 c3b5nF_
not modelled
74.1
13
PDB header: membrane proteinChain: F: PDB Molecule: protein sso1;PDBTitle: structure of the yeast plasma membrane snare complex
82 c3ojaB_
not modelled
73.6
10
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
83 c2qhkA_
not modelled
70.0
15
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of methyl-accepting chemotaxis protein from vibrio2 parahaemolyticus rimd 2210633
84 c1m1jA_
not modelled
64.2
10
PDB header: blood clottingChain: A: PDB Molecule: fibrinogen alpha subunit;PDBTitle: crystal structure of native chicken fibrinogen with two different2 bound ligands
85 d1joya_
not modelled
63.8
15
Fold: ROP-likeSuperfamily: Homodimeric domain of signal transducing histidine kinaseFamily: Homodimeric domain of signal transducing histidine kinase86 c3ipdB_
not modelled
63.7
3
PDB header: exocytosisChain: B: PDB Molecule: syntaxin-1a;PDBTitle: helical extension of the neuronal snare complex into the2 membrane, spacegroup i 21 21 21
87 c2npsB_
not modelled
62.4
14
PDB header: transport proteinChain: B: PDB Molecule: syntaxin 13;PDBTitle: crystal structure of the early endosomal snare complex
88 d3by8a1
not modelled
61.9
23
Fold: Profilin-likeSuperfamily: Sensory domain-likeFamily: Sensory domain of two-component sensor kinase89 c1deqF_
not modelled
58.0
7
PDB header: PDB COMPND: 90 c1n7sB_
not modelled
57.5
3
PDB header: transport proteinChain: B: PDB Molecule: syntaxin 1a;PDBTitle: high resolution structure of a truncated neuronal snare2 complex
91 c3ka5A_
not modelled
56.5
24
PDB header: chaperoneChain: A: PDB Molecule: ribosome-associated protein y (psrp-1);PDBTitle: crystal structure of ribosome-associated protein y (psrp-1)2 from clostridium acetobutylicum. northeast structural3 genomics consortium target id car123a
92 d2p7ja2
not modelled
48.5
10
Fold: Profilin-likeSuperfamily: Sensory domain-likeFamily: YkuI C-terminal domain-like93 c1kmiZ_
not modelled
48.2
13
PDB header: signaling proteinChain: Z: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of an e.coli chemotaxis protein, chez
94 d2c2aa1
not modelled
46.9
14
Fold: ROP-likeSuperfamily: Homodimeric domain of signal transducing histidine kinaseFamily: Homodimeric domain of signal transducing histidine kinase95 c2qltA_
not modelled
43.1
15
PDB header: hydrolaseChain: A: PDB Molecule: (dl)-glycerol-3-phosphatase 1;PDBTitle: crystal structure of an isoform of dl-glycerol-3-phosphatase, rhr2p,2 from saccharomyces cerevisiae
96 c3k2tA_
not modelled
42.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lmo2511 protein;PDBTitle: crystal structure of lmo2511 protein from listeria2 monocytogenes, northeast structural genomics consortium3 target lkr84a
97 c3lifA_
not modelled
40.5
17
PDB header: signaling proteinChain: A: PDB Molecule: putative diguanylate cyclase (ggdef) with pas/pac domain;PDBTitle: crystal structure of the extracellular domain of the putative2 histidine kinase rphk1s-z16
98 c3c25A_
not modelled
34.2
17
PDB header: hydrolase/dnaChain: A: PDB Molecule: noti restriction endonuclease;PDBTitle: crystal structure of noti restriction endonuclease bound to cognate2 dna
99 c2d4yA_
not modelled
31.9
8
PDB header: structural proteinChain: A: PDB Molecule: flagellar hook-associated protein 1;PDBTitle: crystal structure of a 49k fragment of hap1 (flgk)
100 c1s94A_
not modelled
30.0
6
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: s-syntaxin;PDBTitle: crystal structure of the habc domain of neuronal syntaxin from the2 squid loligo pealei
101 d1s94a_
not modelled
30.0
6
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins102 c2d3eD_
not modelled
25.5
8
PDB header: contractile proteinChain: D: PDB Molecule: general control protein gcn4 and tropomyosin 1PDBTitle: crystal structure of the c-terminal fragment of rabbit2 skeletal alpha-tropomyosin
103 c2p7jA_
not modelled
25.0
10
PDB header: transcriptionChain: A: PDB Molecule: putative sensory box/ggdef family protein;PDBTitle: crystal structure of the domain of putative sensory box/ggdef family2 protein from vibrio parahaemolyticus
104 c3u59C_
not modelled
21.8
14
PDB header: contractile proteinChain: C: PDB Molecule: tropomyosin beta chain;PDBTitle: n-terminal 98-aa fragment of smooth muscle tropomyosin beta
105 c2pkeA_
not modelled
21.7
13
PDB header: hydrolaseChain: A: PDB Molecule: haloacid delahogenase-like family hydrolase;PDBTitle: crystal structure of haloacid delahogenase-like family hydrolase2 (np_639141.1) from xanthomonas campestris at 1.81 a resolution
106 c2xhaB_
not modelled
21.4
22
PDB header: transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of domain 2 of thermotoga maritima n-utilization2 substance g (nusg)