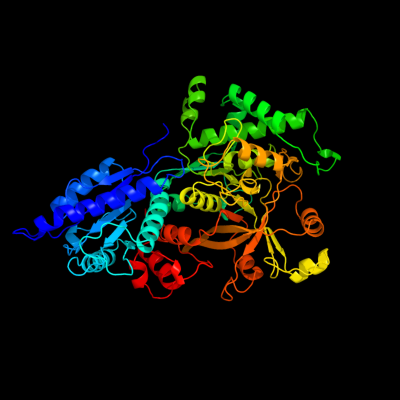
| 1 | c1fuiB_
|
|
 |
100.0 |
100 |
PDB header:isomerase
Chain: B: PDB Molecule:l-fucose isomerase;
PDBTitle: l-fucose isomerase from escherichia coli
|
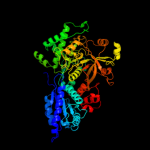
| 2 | c3a9rA_
|
|
 |
100.0 |
65 |
PDB header:isomerase
Chain: A: PDB Molecule:d-arabinose isomerase;
PDBTitle: x-ray structures of bacillus pallidus d-arabinose2 isomerasecomplex with (4r)-2-methylpentane-2,4-diol
|
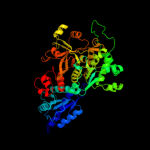
| 3 | d1fuia1
|
|
 |
100.0 |
100 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:FucI/AraA C-terminal domain-like
Family:L-fucose isomerase, C-terminal domain |
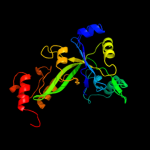
| 4 | d1fuia2
|
|
 |
100.0 |
100 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:L-fucose isomerase, N-terminal and second domains |
| 5 | d2ajta2
|
|
 |
100.0 |
11 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:AraA N-terminal and middle domain-like |
| 6 | c2hxgB_
|
|
 |
100.0 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:l-arabinose isomerase;
PDBTitle: crystal structure of mn2+ bound ecai
|
| 7 | d2ajta1
|
|
 |
83.6 |
19 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:FucI/AraA C-terminal domain-like
Family:AraA C-terminal domain-like |
| 8 | d1kfia1
|
|
 |
83.0 |
15 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
| 9 | d3pmga1
|
|
 |
82.4 |
15 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
| 10 | d1mkza_
|
|
 |
78.6 |
16 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 11 | d1xi8a3
|
|
 |
77.0 |
18 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 12 | c3lftA_
|
|
 |
73.4 |
13 |
PDB header:structure genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the crystal structure of the abc domain in complex with l-trp from2 streptococcus pneumonia to 1.35a
|
| 13 | d2ieaa3
|
|
 |
72.1 |
16 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Transketolase C-terminal domain-like |
| 14 | c3qfeB_
|
|
 |
70.8 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:putative dihydrodipicolinate synthase family protein;
PDBTitle: crystal structures of a putative dihydrodipicolinate synthase family2 protein from coccidioides immitis
|
| 15 | c2p10D_
|
|
 |
69.2 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
| 16 | c2pjkA_
|
|
 |
68.8 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:178aa long hypothetical molybdenum cofactor
PDBTitle: structure of hypothetical molybdenum cofactor biosynthesis2 protein b from sulfolobus tokodaii
|
| 17 | d2p10a1
|
|
 |
67.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Mll9387-like |
| 18 | d1r9da_
|
|
 |
66.2 |
19 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:PFL-like |
| 19 | c3m9yB_
|
|
 |
63.6 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from methicillin2 resistant staphylococcus aureus at 1.9 angstrom resolution
|
| 20 | d2f7wa1
|
|
 |
63.5 |
20 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 21 | c3gjzB_ |
|
not modelled |
63.2 |
15 |
PDB header:immune system
Chain: B: PDB Molecule:microcin immunity protein mccf;
PDBTitle: crystal structure of microcin immunity protein mccf from bacillus2 anthracis str. ames
|
| 22 | d1uuya_ |
|
not modelled |
61.9 |
12 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 23 | d2a5la1 |
|
not modelled |
61.3 |
15 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 24 | d2bfda1 |
|
not modelled |
61.2 |
15 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase PP module |
| 25 | d1zl0a2 |
|
not modelled |
61.0 |
11 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:LD-carboxypeptidase A N-terminal domain-like |
| 26 | c2wc1A_ |
|
not modelled |
60.1 |
18 |
PDB header:electron transport
Chain: A: PDB Molecule:flavodoxin;
PDBTitle: three-dimensional structure of the nitrogen fixation2 flavodoxin (niff) from rhodobacter capsulatus at 2.2 a
|
| 27 | c3s5oA_ |
|
not modelled |
58.6 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase, mitochondrial;
PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
|
| 28 | d1yoba1 |
|
not modelled |
58.4 |
15 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 29 | d1y5ea1 |
|
not modelled |
56.6 |
16 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 30 | d1w3ia_ |
|
not modelled |
56.5 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 31 | c2vlbC_ |
|
not modelled |
55.2 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:arylmalonate decarboxylase;
PDBTitle: structure of unliganded arylmalonate decarboxylase
|
| 32 | d2g2ca1 |
|
not modelled |
54.9 |
23 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 33 | c3snrA_ |
|
not modelled |
53.2 |
8 |
PDB header:transport protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: rpd_1889 protein, an extracellular ligand-binding receptor from2 rhodopseudomonas palustris.
|
| 34 | d1xxxa1 |
|
not modelled |
52.3 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 35 | c2rfgB_ |
|
not modelled |
51.6 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|
| 36 | d1di6a_ |
|
not modelled |
50.8 |
21 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 37 | c3ip5A_ |
|
not modelled |
50.8 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter, substrate binding protein (amino acid);
PDBTitle: structure of atu2422-gaba receptor in complex with alanine
|
| 38 | c3h6hB_ |
|
not modelled |
49.4 |
11 |
PDB header:membrane protein
Chain: B: PDB Molecule:glutamate receptor, ionotropic kainate 2;
PDBTitle: crystal structure of the glur6 amino terminal domain dimer assembly2 mpd form
|
| 39 | d1m6ja_ |
|
not modelled |
49.3 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 40 | c2zkiH_ |
|
not modelled |
48.3 |
15 |
PDB header:transcription
Chain: H: PDB Molecule:199aa long hypothetical trp repressor binding
PDBTitle: crystal structure of hypothetical trp repressor binding2 protein from sul folobus tokodaii (st0872)
|
| 41 | c1yyaA_ |
|
not modelled |
46.8 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of tt0473, putative triosephosphate isomerase from2 thermus thermophilus hb8
|
| 42 | d2p12a1 |
|
not modelled |
46.2 |
26 |
Fold:FomD barrel-like
Superfamily:FomD-like
Family:FomD-like |
| 43 | c2dp3A_ |
|
not modelled |
45.1 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of a double mutant (c202a/a198v) of triosephosphate2 isomerase from giardia lamblia
|
| 44 | c2y8nC_ |
|
not modelled |
44.9 |
20 |
PDB header:lyase
Chain: C: PDB Molecule:4-hydroxyphenylacetate decarboxylase large subunit;
PDBTitle: crystal structure of glycyl radical enzyme
|
| 45 | d1b9ba_ |
|
not modelled |
44.2 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 46 | d1o5xa_ |
|
not modelled |
43.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 47 | d1dkia_ |
|
not modelled |
42.2 |
24 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Papain-like |
| 48 | c3ksmA_ |
|
not modelled |
42.2 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:abc-type sugar transport system, periplasmic component;
PDBTitle: crystal structure of abc-type sugar transport system, periplasmic2 component from hahella chejuensis
|
| 49 | d1r2ra_ |
|
not modelled |
41.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 50 | c3q4nA_ |
|
not modelled |
40.4 |
29 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein mj0754;
PDBTitle: crystal structure of hypothetical protein mj0754 from methanococcus2 jannaschii dsm 2661
|
| 51 | d1aw1a_ |
|
not modelled |
40.4 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 52 | d1pvja_ |
|
not modelled |
40.1 |
23 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Papain-like |
| 53 | c3bb7A_ |
|
not modelled |
39.9 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:interpain a;
PDBTitle: structure of prevotella intermedia prointerpain a fragment 39-3592 (mutant c154a)
|
| 54 | c2r8wB_ |
|
not modelled |
39.3 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:agr_c_1641p;
PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
|
| 55 | d1n55a_ |
|
not modelled |
39.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 56 | c3gr7A_ |
|
not modelled |
38.6 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal2 crystal form
|
| 57 | d1suxa_ |
|
not modelled |
38.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 58 | c3i45A_ |
|
not modelled |
37.7 |
11 |
PDB header:signaling protein
Chain: A: PDB Molecule:twin-arginine translocation pathway signal protein;
PDBTitle: crystal structure of putative twin-arginine translocation pathway2 signal protein from rhodospirillum rubrum atcc 11170
|
| 59 | c2qh8A_ |
|
not modelled |
37.5 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of conserved domain protein from vibrio2 cholerae o1 biovar eltor str. n16961
|
| 60 | d1neya_ |
|
not modelled |
37.1 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 61 | c2qvcC_ |
|
not modelled |
36.9 |
14 |
PDB header:transport protein
Chain: C: PDB Molecule:sugar abc transporter, periplasmic sugar-binding
PDBTitle: crystal structure of a periplasmic sugar abc transporter2 from thermotoga maritima
|
| 62 | d1qo0a_ |
|
not modelled |
36.9 |
14 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 63 | c3td9A_ |
|
not modelled |
36.5 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:branched chain amino acid abc transporter, periplasmic
PDBTitle: crystal structure of a leucine binding protein livk (tm1135) from2 thermotoga maritima msb8 at 1.90 a resolution
|
| 64 | c3bbaB_ |
|
not modelled |
36.3 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:interpain a;
PDBTitle: structure of active wild-type prevotella intermedia interpain a2 cysteine protease
|
| 65 | d1tjya_ |
|
not modelled |
36.2 |
13 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 66 | d1trea_ |
|
not modelled |
35.4 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 67 | d1ycga1 |
|
not modelled |
35.4 |
12 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 68 | c3b4uB_ |
|
not modelled |
35.1 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from agrobacterium2 tumefaciens str. c58
|
| 69 | c3krsB_ |
|
not modelled |
34.7 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: structure of triosephosphate isomerase from cryptosporidium parvum at2 1.55a resolution
|
| 70 | d2auna2 |
|
not modelled |
33.9 |
11 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:LD-carboxypeptidase A N-terminal domain-like |
| 71 | c3kxqB_ |
|
not modelled |
33.5 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from bartonella2 henselae at 1.6a resolution
|
| 72 | d2ftsa3 |
|
not modelled |
33.4 |
9 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 73 | c3na8A_ |
|
not modelled |
32.9 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
|
| 74 | c3qi7A_ |
|
not modelled |
32.5 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of a putative transcriptional regulator2 (yp_001089212.1) from clostridium difficile 630 at 1.86 a resolution
|
| 75 | c1zrsB_ |
|
not modelled |
31.8 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: wild-type ld-carboxypeptidase
|
| 76 | c3g0sA_ |
|
not modelled |
31.8 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from salmonella typhimurium lt2
|
| 77 | c3i09A_ |
|
not modelled |
31.4 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic branched-chain amino acid-binding protein;
PDBTitle: crystal structure of a periplasmic binding protein (bma2936) from2 burkholderia mallei at 1.80 a resolution
|
| 78 | c2x7xA_ |
|
not modelled |
31.1 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: fructose binding periplasmic domain of hybrid two component2 system bt1754
|
| 79 | c3daqB_ |
|
not modelled |
30.6 |
7 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
| 80 | c3gvgA_ |
|
not modelled |
30.6 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from mycobacterium2 tuberculosis
|
| 81 | c3n5lA_ |
|
not modelled |
30.4 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:binding protein component of abc phosphonate transporter;
PDBTitle: crystal structure of a binding protein component of abc phosphonate2 transporter (pa3383) from pseudomonas aeruginosa at 1.97 a resolution
|
| 82 | c3noeA_ |
|
not modelled |
30.1 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
| 83 | c3mjdA_ |
|
not modelled |
29.8 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:orotate phosphoribosyltransferase;
PDBTitle: 1.9 angstrom crystal structure of orotate2 phosphoribosyltransferase (pyre) francisella tularensis.
|
| 84 | d1kv5a_ |
|
not modelled |
29.7 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 85 | d1qs0a_ |
|
not modelled |
29.6 |
13 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase PP module |
| 86 | c3fluD_ |
|
not modelled |
29.6 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 87 | c3r85H_ |
|
not modelled |
29.3 |
60 |
PDB header:apoptosis
Chain: H: PDB Molecule:heme-binding protein 2;
PDBTitle: crystal structure of human soul bh3 domain in complex with bcl-xl
|
| 88 | d1o5ka_ |
|
not modelled |
29.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 89 | c3cprB_ |
|
not modelled |
29.0 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthetase;
PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
|
| 90 | d1jpma1 |
|
not modelled |
28.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 91 | c3o3nA_ |
|
not modelled |
28.6 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:alpha-subunit 2-hydroxyisocaproyl-coa dehydratase;
PDBTitle: (r)-2-hydroxyisocaproyl-coa dehydratase in complex with its substrate2 (r)-2-hydroxyisocaproyl-coa
|
| 92 | c2l9dA_ |
|
not modelled |
28.6 |
42 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of the protein yp_546394.1, the first structural2 representative of the pfam family pf12112
|
| 93 | d1mo0a_ |
|
not modelled |
28.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 94 | c2vc6A_ |
|
not modelled |
28.3 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of mosa from s. meliloti with pyruvate bound
|
| 95 | d2liva_ |
|
not modelled |
28.3 |
10 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
| 96 | c3th6B_ |
|
not modelled |
28.2 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from rhipicephalus2 (boophilus) microplus.
|
| 97 | d2a6na1 |
|
not modelled |
28.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 98 | c2yxgD_ |
|
not modelled |
27.7 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
| 99 | c2r94B_ |
|
not modelled |
27.6 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxy-(6-phospho-)gluconate aldolase;
PDBTitle: crystal structure of kd(p)ga from t.tenax
|
| 100 | d1sw3a_ |
|
not modelled |
27.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 101 | c3r85E_ |
|
not modelled |
27.2 |
60 |
PDB header:apoptosis
Chain: E: PDB Molecule:heme-binding protein 2;
PDBTitle: crystal structure of human soul bh3 domain in complex with bcl-xl
|
| 102 | c3r85G_ |
|
not modelled |
27.0 |
60 |
PDB header:apoptosis
Chain: G: PDB Molecule:heme-binding protein 2;
PDBTitle: crystal structure of human soul bh3 domain in complex with bcl-xl
|
| 103 | d2arka1 |
|
not modelled |
26.9 |
19 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 104 | c3lerA_ |
|
not modelled |
26.7 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 105 | c3sg0A_ |
|
not modelled |
26.5 |
17 |
PDB header:signaling protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: the crystal structure of an extracellular ligand-binding receptor from2 rhodopseudomonas palustris haa2
|
| 106 | d1uz5a3 |
|
not modelled |
26.2 |
12 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 107 | c3eb2A_ |
|
not modelled |
25.9 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 108 | c3n0xA_ |
|
not modelled |
25.7 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:possible substrate binding protein of abc transporter
PDBTitle: crystal structure of an abc-type branched-chain amino acid transporter2 (rpa4397) from rhodopseudomonas palustris cga009 at 1.50 a resolution
|
| 109 | d1vmea1 |
|
not modelled |
25.5 |
6 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 110 | c2p1nD_ |
|
not modelled |
24.8 |
16 |
PDB header:signaling protein
Chain: D: PDB Molecule:skp1-like protein 1a;
PDBTitle: mechanism of auxin perception by the tir1 ubiqutin ligase
|
| 111 | c2uuvC_ |
|
not modelled |
24.7 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:alkyldihydroxyacetonephosphate synthase;
PDBTitle: alkyldihydroxyacetonephosphate synthase in p1
|
| 112 | d2btma_ |
|
not modelled |
24.5 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 113 | c3pueA_ |
|
not modelled |
24.3 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
| 114 | d1f74a_ |
|
not modelled |
24.2 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 115 | c3eo5A_ |
|
not modelled |
24.0 |
36 |
PDB header:cell adhesion
Chain: A: PDB Molecule:resuscitation-promoting factor rpfb;
PDBTitle: crystal structure of the resuscitation promoting factor rpfb
|
| 116 | c2v9dB_ |
|
not modelled |
23.6 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
|
| 117 | c3n2lA_ |
|
not modelled |
23.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:orotate phosphoribosyltransferase;
PDBTitle: 2.1 angstrom resolution crystal structure of an orotate2 phosphoribosyltransferase (pyre) from vibrio cholerae o1 biovar eltor3 str. n16961
|
| 118 | c3n2xB_ |
|
not modelled |
22.8 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:uncharacterized protein yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
|
| 119 | c3g23A_ |
|
not modelled |
22.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:ld-carboxypeptidase a;
PDBTitle: crystal structure of a ld-carboxypeptidase a (saro_1426) from2 novosphingobium aromaticivorans dsm at 1.89 a resolution
|
| 120 | d2dara1 |
|
not modelled |
22.3 |
40 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |