1 c2htbB_
100.0
83
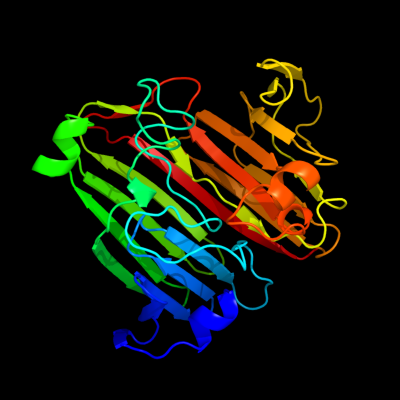
PDB header: isomeraseChain: B: PDB Molecule: putative enzyme related to aldose 1-epimerase;PDBTitle: crystal structure of a putative mutarotase (yead) from2 salmonella typhimurium in monoclinic form
2 d1jova_
100.0
35
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Hypothetical protein HI13173 c2cisA_
100.0
28
PDB header: isomeraseChain: A: PDB Molecule: hexose-6-phosphate mutarotase;PDBTitle: structure-based functional annotation: yeast ymr099c codes2 for a d-hexose-6-phosphate mutarotase. complex with3 tagatose-6-phosphate
4 c3k25B_
100.0
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: slr1438 protein;PDBTitle: crystal structure of slr1438 protein from synechocystis sp. pcc 6803,2 northeast structural genomics consortium target sgr112
5 c3dcdA_
100.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: galactose mutarotase related enzyme;PDBTitle: x-ray structure of the galactose mutarotase related enzyme q5fkd7 from2 lactobacillus acidophilus at the resolution 1.9a. northeast3 structural genomics consortium target lar33.
6 c3os7B_
100.0
19
PDB header: isomeraseChain: B: PDB Molecule: galactose mutarotase-like protein;PDBTitle: crystal structure of a galactose mutarotase-like protein (ca_c0697)2 from clostridium acetobutylicum at 1.80 a resolution
7 c3q1nA_
100.0
21
PDB header: isomeraseChain: A: PDB Molecule: galactose mutarotase related enzyme;PDBTitle: crystal structure of a galactose mutarotase-like protein (lsei_2598)2 from lactobacillus casei atcc 334 at 1.61 a resolution
8 c3os7D_
100.0
19
PDB header: isomeraseChain: D: PDB Molecule: galactose mutarotase-like protein;PDBTitle: crystal structure of a galactose mutarotase-like protein (ca_c0697)2 from clostridium acetobutylicum at 1.80 a resolution
9 c3nreB_
100.0
17
PDB header: isomeraseChain: B: PDB Molecule: aldose 1-epimerase;PDBTitle: crystal structure of a putative aldose 1-epimerase (b2544) from2 escherichia coli k12 at 1.59 a resolution
10 c3mwxA_
100.0
15
PDB header: isomeraseChain: A: PDB Molecule: aldose 1-epimerase;PDBTitle: crystal structure of a putative galactose mutarotase (bsu18360) from2 bacillus subtilis at 1.45 a resolution
11 c3imhB_
100.0
16
PDB header: isomeraseChain: B: PDB Molecule: galactose-1-epimerase;PDBTitle: crystal structure of galactose 1-epimerase from lactobacillus2 acidophilus ncfm
12 d1nsza_
100.0
17
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)13 d1lura_
100.0
15
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)14 c1ygaA_
100.0
13
PDB header: isomeraseChain: A: PDB Molecule: hypothetical 37.9 kda protein in bio3-hxt17PDBTitle: crystal structure of saccharomyces cerevisiae yn9a protein,2 new york structural genomics consortium
15 d1so0a_
100.0
13
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)16 d1z45a1
100.0
14
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)17 c1z45A_
100.0
15
PDB header: isomeraseChain: A: PDB Molecule: gal10 bifunctional protein;PDBTitle: crystal structure of the gal10 fusion protein galactose2 mutarotase/udp-galactose 4-epimerase from saccharomyces3 cerevisiae complexed with nad, udp-glucose, and galactose
18 c3ty1B_
99.9
14
PDB header: isomeraseChain: B: PDB Molecule: hypothetical aldose 1-epimerase;PDBTitle: crystal structure of a hypothetical aldose 1-epimerase (kpn_04629)2 from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 1.90 a3 resolution
19 c3bs6B_
96.6
11
PDB header: membrane protein, protein transportChain: B: PDB Molecule: inner membrane protein oxaa;PDBTitle: 1.8 angstrom crystal structure of the periplasmic domain of2 the membrane insertase yidc
20 c3blcB_
96.4
10
PDB header: chaperone,protein transportChain: B: PDB Molecule: inner membrane protein oxaa;PDBTitle: crystal structure of the periplasmic domain of the escherichia coli2 yidc
21 d1ejxb_
not modelled
37.9
10
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit22 d4ubpb_
not modelled
36.4
14
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit23 c2yfnA_
not modelled
35.5
11
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase-sucrose kinase agask;PDBTitle: galactosidase domain of alpha-galactosidase-sucrose kinase,2 agask
24 d1e9ya1
not modelled
35.3
10
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit25 d1jz8a4
not modelled
32.7
15
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: beta-Galactosidase, domain 526 c2js4A_
not modelled
31.2
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0434 protein bb2007;PDBTitle: solution nmr structure of bordetella bronchiseptica protein2 bb2007. northeast structural genomics consortium target3 bor54
27 c3qgaD_
not modelled
29.4
10
PDB header: hydrolaseChain: D: PDB Molecule: fusion of urease beta and gamma subunits;PDBTitle: 3.0 a model of iron containing urease urea2b2 from helicobacter2 mustelae
28 c1e9zA_
not modelled
26.7
10
PDB header: hydrolaseChain: A: PDB Molecule: urease subunit alpha;PDBTitle: crystal structure of helicobacter pylori urease
29 d1fcqa_
not modelled
21.2
32
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Bee venom hyaluronidase30 c2jr6A_
not modelled
18.6
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0434 protein nma0874;PDBTitle: solution structure of upf0434 protein nma0874. northeast structural2 genomics target mr32
31 d2pk7a1
not modelled
17.7
45
Fold: Trm112p-likeSuperfamily: Trm112p-likeFamily: Trm112p-like32 d2jnya1
not modelled
16.8
36
Fold: Trm112p-likeSuperfamily: Trm112p-likeFamily: Trm112p-like33 d2hf1a1
not modelled
16.6
27
Fold: Trm112p-likeSuperfamily: Trm112p-likeFamily: Trm112p-like34 c2k5rA_
not modelled
14.6
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein xf2673;PDBTitle: solution nmr structure of xf2673 from xylella fastidiosa.2 northeast structural genomics consortium target xfr39
35 c2kpiA_
not modelled
14.3
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein sco3027;PDBTitle: solution nmr structure of streptomyces coelicolor sco30272 modeled with zn+2 bound, northeast structural genomics3 consortium target rr58
36 d1pxva_
not modelled
11.4
29
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Papain-like37 c1y4hA_
not modelled
9.8
29
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: cysteine protease;PDBTitle: wild type staphopain-staphostatin complex
38 c1h4tD_
not modelled
9.0
16
PDB header: aminoacyl-trna synthetaseChain: D: PDB Molecule: prolyl-trna synthetase;PDBTitle: prolyl-trna synthetase from thermus thermophilus complexed2 with l-proline
39 c1qf6A_
not modelled
8.9
25
PDB header: ligase/rnaChain: A: PDB Molecule: threonyl-trna synthetase;PDBTitle: structure of e. coli threonyl-trna synthetase complexed with its2 cognate trna
40 c2qeaB_
not modelled
8.4
10
PDB header: oxidoreductaseChain: B: PDB Molecule: putative general stress protein 26;PDBTitle: crystal structure of a putative general stress protein 26 (jann_0955)2 from jannaschia sp. ccs1 at 2.46 a resolution
41 c1nj8C_
not modelled
8.3
8
PDB header: ligaseChain: C: PDB Molecule: proline-trna synthetase;PDBTitle: crystal structure of prolyl-trna synthetase from2 methanocaldococcus janaschii
42 d1pgw21
not modelled
7.8
21
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP43 d1ny721
not modelled
7.4
26
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP44 d1pgl21
not modelled
7.3
21
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP45 c3qbtH_
not modelled
7.2
7
PDB header: protein transport/hydrolaseChain: H: PDB Molecule: inositol polyphosphate 5-phosphatase ocrl-1;PDBTitle: crystal structure of ocrl1 540-678 in complex with rab8a:gppnhp
46 c3dmbA_
not modelled
7.1
24
PDB header: oxidoreductaseChain: A: PDB Molecule: putative general stress protein 26 with a pnp-oxidase likePDBTitle: crystal structure of a putative general stress family protein2 (xcc2264) from xanthomonas campestris pv. campestris at 2.30 a3 resolution
47 c1yewI_
not modelled
6.6
5
PDB header: oxidoreductase, membrane proteinChain: I: PDB Molecule: particulate methane monooxygenase, b subunit;PDBTitle: crystal structure of particulate methane monooxygenase
48 c3rgbA_
not modelled
6.6
5
PDB header: oxidoreductaseChain: A: PDB Molecule: methane monooxygenase subunit b2;PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
49 d1k1xa2
not modelled
6.6
15
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: 4-alpha-glucanotransferase, C-terminal domain50 c3ialB_
not modelled
6.6
9
PDB header: ligaseChain: B: PDB Molecule: prolyl-trna synthetase;PDBTitle: giardia lamblia prolyl-trna synthetase in complex with prolyl-2 adenylate
51 d1th5a1
not modelled
6.4
21
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like52 c3bmzA_
not modelled
5.1
22
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: violacein biosynthetic enzyme vioe
53 c2r5iL_
not modelled
5.1
26
PDB header: viral proteinChain: L: PDB Molecule: l1 protein;PDBTitle: pentamer structure of major capsid protein l1 of human2 papilloma virus type 18