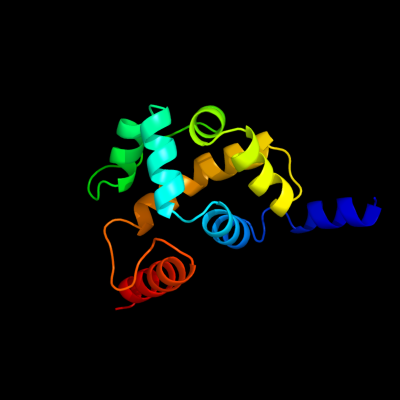
1 d1z67a1
100.0
97
Fold: YidB-likeSuperfamily: YidB-likeFamily: YidB-like2 d1b8za_
68.4
21
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein3 d1huua_
66.2
18
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein4 c2lfcA_
62.9
4
PDB header: oxidoreductaseChain: A: PDB Molecule: fumarate reductase, flavoprotein subunit;PDBTitle: solution nmr structure of fumarate reductase flavoprotein subunit from2 lactobacillus plantarum, northeast structural genomics consortium3 target lpr145j
5 d1y0pa3
62.4
21
Fold: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainSuperfamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain6 d1d4ca3
57.5
8
Fold: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainSuperfamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain7 c1tr8A_
57.3
15
PDB header: chaperoneChain: A: PDB Molecule: conserved protein (mth177);PDBTitle: crystal structure of archaeal nascent polypeptide-associated complex2 (aenac)
8 d1mula_
53.3
21
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein9 d1p71a_
51.6
28
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein10 c3c4iA_
49.5
26
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding protein hu homolog;PDBTitle: crystal structure analysis of n terminal region containing the2 dimerization domain and dna binding domain of hu protein(histone like3 protein-dna binding) from mycobacterium tuberculosis [h37rv]
11 d1qo8a3
47.6
13
Fold: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainSuperfamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain12 d1tafb_
47.3
25
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs13 d1owfa_
43.0
26
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein14 d1l0oc_
41.6
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain15 c1l0oC_
41.6
17
PDB header: protein bindingChain: C: PDB Molecule: sigma factor;PDBTitle: crystal structure of the bacillus stearothermophilus anti-2 sigma factor spoiiab with the sporulation sigma factor3 sigmaf
16 d1exea_
40.2
21
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein17 d2o97b1
40.1
11
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein18 c3ebnD_
37.5
20
PDB header: hydrolaseChain: D: PDB Molecule: replicase polyprotein 1ab;PDBTitle: a special dimerization of sars-cov main protease c-terminal2 domain due to domain-swapping
19 d1uxca_
34.8
25
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator20 d2p7vb1
34.6
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain21 d2hsga1
not modelled
33.6
20
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator22 d2phcb1
not modelled
33.1
13
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: PH0987 C-terminal domain-like23 d1qpza1
not modelled
32.1
25
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator24 d1lcda_
not modelled
32.1
25
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator25 c2iifA_
not modelled
31.8
26
PDB header: recombination/dnaChain: A: PDB Molecule: integration host factor;PDBTitle: single chain integration host factor mutant protein (scihf2-2 k45ae) in complex with dna
26 d2duca1
not modelled
30.3
20
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold27 c2zp2B_
not modelled
30.2
4
PDB header: transferase inhibitorChain: B: PDB Molecule: kinase a inhibitor;PDBTitle: c-terminal domain of kipi from bacillus subtilis
28 c2w7nA_
not modelled
30.0
18
PDB header: transcription/dnaChain: A: PDB Molecule: trfb transcriptional repressor protein;PDBTitle: crystal structure of kora bound to operator dna: insight2 into repressor cooperation in rp4 gene regulation
29 c2lcvA_
not modelled
28.4
35
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional repressor cytr;PDBTitle: structure of the cytidine repressor dna-binding domain; an alternate2 calculation
30 d2cg4a1
not modelled
26.8
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain31 d1uxda_
not modelled
25.8
25
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator32 d2bjca1
not modelled
25.8
25
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator33 c2l8nA_
not modelled
23.9
35
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional repressor cytr;PDBTitle: nmr structure of the cytidine repressor dna binding domain in presence2 of operator half-site dna
34 d1i1ga1
not modelled
23.7
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain35 c3h5tA_
not modelled
23.4
25
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, laci family;PDBTitle: crystal structure of a transcriptional regulator, lacl2 family protein from corynebacterium glutamicum
36 c1bdhA_
not modelled
22.5
25
PDB header: transcription/dnaChain: A: PDB Molecule: protein (purine repressor);PDBTitle: purine repressor mutant-hypoxanthine-palindromic operator2 complex
37 d1z6ra1
not modelled
21.9
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ROK associated domain38 d1ku7a_
not modelled
21.4
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain39 d1efaa1
not modelled
21.1
25
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator40 d2cyya1
not modelled
21.1
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain41 d2fug21
not modelled
21.1
4
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: NQO2-like42 d1ku3a_
not modelled
19.7
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain43 c3kjxD_
not modelled
19.1
30
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator, laci family;PDBTitle: crystal structure of a transcriptional regulator, lacl2 family protein from silicibacter pomeroyi
44 d1wh5a_
not modelled
19.0
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain45 c1or7A_
not modelled
19.0
13
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma-e factor;PDBTitle: crystal structure of escherichia coli sigmae with the cytoplasmic2 domain of its anti-sigma rsea
46 c2phcB_
not modelled
18.5
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein ph0987;PDBTitle: crystal structure of conserved uncharacterized protein ph0987 from2 pyrococcus horikoshii
47 d1tw3a1
not modelled
17.7
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Plant O-methyltransferase, N-terminal domain48 c1zvvA_
not modelled
17.7
19
PDB header: transcription/dnaChain: A: PDB Molecule: glucose-resistance amylase regulator;PDBTitle: crystal structure of a ccpa-crh-dna complex
49 d1ttya_
not modelled
17.4
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain50 d1qzza1
not modelled
17.3
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Plant O-methyltransferase, N-terminal domain51 d3proc1
not modelled
17.2
13
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Alpha-lytic protease prodomainFamily: Alpha-lytic protease prodomain52 d1x2na1
not modelled
16.8
0
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain53 d2gqba1
not modelled
16.7
20
Fold: RPA2825-likeSuperfamily: RPA2825-likeFamily: RPA2825-like54 d3e11a1
not modelled
16.7
11
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: TTHA0227-like55 d1wi9a_
not modelled
16.5
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PCI domain (PINT motif)56 d2cfxa1
not modelled
16.4
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain57 c2da4A_
not modelled
15.6
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein dkfzp686k21156;PDBTitle: solution structure of the homeobox domain of the2 hypothetical protein, dkfzp686k21156
58 d1e3oc1
not modelled
15.5
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain59 c3d23A_
not modelled
15.4
15
PDB header: hydrolaseChain: A: PDB Molecule: 3c-like proteinase;PDBTitle: main protease of hcov-hku1
60 d1x2ma1
not modelled
15.0
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain61 d1or7a1
not modelled
15.0
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain62 c2kz3A_
not modelled
14.5
19
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein rad51l3;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for human rad51d2 from 1 to 83
63 c2cg4B_
not modelled
14.3
17
PDB header: transcriptionChain: B: PDB Molecule: regulatory protein asnc;PDBTitle: structure of e.coli asnc
64 c3majA_
not modelled
14.3
15
PDB header: dna binding proteinChain: A: PDB Molecule: dna processing chain a;PDBTitle: crystal structure of putative dna processing protein dpra from2 rhodopseudomonas palustris cga009
65 c2q6fB_
not modelled
14.2
19
PDB header: hydrolaseChain: B: PDB Molecule: infectious bronchitis virus (ibv) main protease;PDBTitle: crystal structure of infectious bronchitis virus (ibv) main2 protease in complex with a michael acceptor inhibitor n3
66 c1rr7A_
not modelled
14.1
18
PDB header: transcriptionChain: A: PDB Molecule: middle operon regulator;PDBTitle: crystal structure of the middle operon regulator protein of2 bacteriophage mu
67 d1rr7a_
not modelled
14.1
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Middle operon regulator, Mor68 d1alna1
not modelled
14.0
29
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase69 c1y66D_
not modelled
13.8
17
PDB header: de novo proteinChain: D: PDB Molecule: engrailed homeodomain;PDBTitle: dioxane contributes to the altered conformation and2 oligomerization state of a designed engrailed homeodomain3 variant
70 c2l4aA_
not modelled
13.8
10
PDB header: dna binding proteinChain: A: PDB Molecule: leucine responsive regulatory protein;PDBTitle: nmr structure of the dna-binding domain of e.coli lrp
71 d1e0ga_
not modelled
13.6
15
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain72 d1bw5a_
not modelled
13.5
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain73 d2cqxa1
not modelled
13.4
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain74 c2yskA_
not modelled
13.4
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ttha1432;PDBTitle: crystal structure of a hypothetical protein ttha1432 from thermus2 thermophilus
75 c2np2B_
not modelled
13.3
10
PDB header: dna binding protein/dnaChain: B: PDB Molecule: hbb;PDBTitle: hbb-dna complex
76 c2qufB_
not modelled
13.2
16
PDB header: transcriptionChain: B: PDB Molecule: transcription factor pf0095;PDBTitle: crystal structure of transcription factor axxa-pf0095 from pyrococcus2 furiosus
77 d1p9sa_
not modelled
13.2
27
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold78 c3mmlD_
not modelled
13.0
4
PDB header: hydrolaseChain: D: PDB Molecule: allophanate hydrolase subunit 1;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
79 d2ozua1
not modelled
12.8
15
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT80 c1alnA_
not modelled
12.7
25
PDB header: hydrolaseChain: A: PDB Molecule: cytidine deaminase;PDBTitle: crystal structure of cytidine deaminase complexed with 3-deazacytidine
81 c2l0kA_
not modelled
12.5
37
PDB header: transcriptionChain: A: PDB Molecule: stage iii sporulation protein d;PDBTitle: nmr solution structure of a transcription factor spoiiid in complex2 with dna
82 c2e1cA_
not modelled
12.5
14
PDB header: transcription/dnaChain: A: PDB Molecule: putative hth-type transcriptional regulator ph1519;PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
83 c3t72o_
not modelled
12.5
19
PDB header: transcription/dnaChain: O: PDB Molecule: pho box dna (strand 1);PDBTitle: phob(e)-sigma70(4)-(rnap-betha-flap-tip-helix)-dna transcription2 activation sub-complex
84 c2da5A_
not modelled
12.4
6
PDB header: dna binding proteinChain: A: PDB Molecule: zinc fingers and homeoboxes protein 3;PDBTitle: solution structure of the second homeobox domain of zinc2 fingers and homeoboxes protein 3 (triple homeobox 13 protein)
85 d1mh3a1
not modelled
12.4
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain86 c2djpA_
not modelled
12.3
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein sb145;PDBTitle: the solution structure of the lysm domain of human2 hypothetical protein sb145
87 d1s7ea1
not modelled
12.2
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain88 d2r5yb1
not modelled
12.2
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain89 c4proD_
not modelled
12.0
12
PDB header: serine proteaseChain: D: PDB Molecule: alpha-lytic protease;PDBTitle: alpha-lytic protease complexed with pro region
90 c2da7A_
not modelled
11.9
6
PDB header: dna binding proteinChain: A: PDB Molecule: zinc finger homeobox protein 1b;PDBTitle: solution structure of the homeobox domain of zinc finger2 homeobox protein 1b (smad interacting protein 1)
91 c3nauA_
not modelled
11.8
22
PDB header: transcriptionChain: A: PDB Molecule: zinc fingers and homeoboxes protein 2;PDBTitle: crystal structure of zhx2 hd2 (zinc-fingers and homeoboxes protein 2,2 homeodomain 2)
92 c3f6wE_
not modelled
11.7
24
PDB header: dna binding proteinChain: E: PDB Molecule: xre-family like protein;PDBTitle: xre-family like protein from pseudomonas syringae pv. tomato str.2 dc3000
93 d1k61a_
not modelled
11.6
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain94 c2a6eF_
not modelled
11.6
20
PDB header: transferaseChain: F: PDB Molecule: rna polymerase sigma factor rpod;PDBTitle: crystal structure of the t. thermophilus rna polymerase2 holoenzyme
95 d2ecba1
not modelled
11.5
5
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain96 d2hddb_
not modelled
11.5
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain97 d1du6a_
not modelled
11.5
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain98 d1j5ya1
not modelled
11.5
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Biotin repressor-like99 c2k5eA_
not modelled
11.4
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of putative uncharacterized protein2 gsu1278 from methanocaldococcus jannaschii, northeast3 structural genomics consortium (nesg) target gsr195