1 c2j5uB_
100.0
28
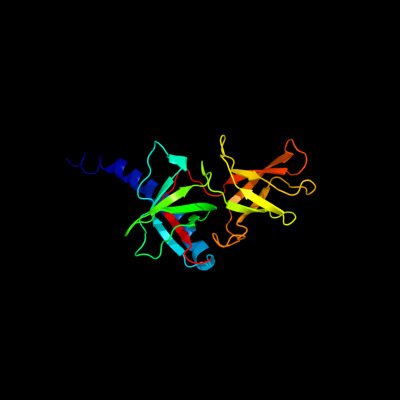
PDB header: cell shape regulationChain: B: PDB Molecule: mrec protein;PDBTitle: mrec lysteria monocytogenes
2 c2qf4A_
100.0
23
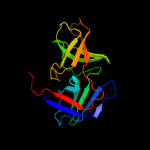
PDB header: structural proteinChain: A: PDB Molecule: cell shape determining protein mrec;PDBTitle: high resolution structure of the major periplasmic domain from the2 cell shape-determining filament mrec (orthorhombic form)
3 c2wvrB_
89.5
26
PDB header: replicationChain: B: PDB Molecule: geminin;PDBTitle: human cdt1:geminin complex
4 c2zxxA_
89.0
28
PDB header: cell cycle/replicationChain: A: PDB Molecule: geminin;PDBTitle: crystal structure of cdt1/geminin complex
5 c1t6fA_
86.3
31
PDB header: cell cycleChain: A: PDB Molecule: geminin;PDBTitle: crystal structure of the coiled-coil dimerization motif of2 geminin
6 c3m9bK_
85.9
18
PDB header: chaperoneChain: K: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
7 c1ci6B_
81.3
25
PDB header: transcriptionChain: B: PDB Molecule: transcription factor c/ebp beta;PDBTitle: transcription factor atf4-c/ebp beta bzip heterodimer
8 d2jdia2
81.0
14
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase9 c1dipA_
80.7
24
PDB header: acetylationChain: A: PDB Molecule: delta-sleep-inducing peptide immunoreactivePDBTitle: the solution structure of porcine delta-sleep-inducing2 peptide immunoreactive peptide, nmr, 10 structures
10 d1fxkc_
80.6
11
Fold: Long alpha-hairpinSuperfamily: PrefoldinFamily: Prefoldin11 d1fx0a2
80.4
14
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase12 d1skyb2
77.3
20
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase13 c2xzrA_
76.4
16
PDB header: cell adhesionChain: A: PDB Molecule: immunoglobulin-binding protein eibd;PDBTitle: escherichia coli immunoglobulin-binding protein eibd 391-438 fused2 to gcn4 adaptors
14 c2jeeA_
74.6
22
PDB header: cell cycleChain: A: PDB Molecule: yiiu;PDBTitle: xray structure of e. coli yiiu
15 c2yy0D_
71.1
32
PDB header: transcriptionChain: D: PDB Molecule: c-myc-binding protein;PDBTitle: crystal structure of ms0802, c-myc-1 binding protein domain2 from homo sapiens
16 d1maba2
70.6
14
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase17 c2oqqB_
70.1
28
PDB header: transcriptionChain: B: PDB Molecule: transcription factor hy5;PDBTitle: crystal structure of hy5 leucine zipper homodimer from2 arabidopsis thaliana
18 c1debA_
69.8
33
PDB header: structural proteinChain: A: PDB Molecule: adenomatous polyposis coli protein;PDBTitle: crystal structure of the n-terminal coiled coil domain from2 apc
19 c3pv4A_
67.0
15
PDB header: hydrolaseChain: A: PDB Molecule: degq;PDBTitle: structure of legionella fallonii degq (delta-pdz2 variant)
20 c3mudA_
66.1
19
PDB header: contractile proteinChain: A: PDB Molecule: dna repair protein xrcc4, tropomyosin alpha-1 chain;PDBTitle: structure of the tropomyosin overlap complex from chicken smooth2 muscle
21 c1ce0B_
not modelled
60.5
25
PDB header: hiv-1 envelope proteinChain: B: PDB Molecule: protein (leucine zipper model h38-p1);PDBTitle: trimerization specificity in hiv-1 gp41: analysis with a2 gcn4 leucine zipper model
22 c1t2kD_
not modelled
59.3
24
PDB header: transcription/dnaChain: D: PDB Molecule: cyclic-amp-dependent transcription factor atf-2;PDBTitle: structure of the dna binding domains of irf3, atf-2 and jun2 bound to dna
23 c2e43A_
not modelled
58.4
19
PDB header: transcription/dnaChain: A: PDB Molecule: ccaat/enhancer-binding protein beta;PDBTitle: crystal structure of c/ebpbeta bzip homodimer k269a mutant2 bound to a high affinity dna fragment
24 c2wukD_
not modelled
58.2
3
PDB header: cell cycleChain: D: PDB Molecule: septum site-determining protein diviva;PDBTitle: diviva n-terminal domain, f17a mutant
25 c3qh9A_
not modelled
58.2
21
PDB header: structural proteinChain: A: PDB Molecule: liprin-beta-2;PDBTitle: human liprin-beta2 coiled-coil
26 c1fosE_
not modelled
57.7
24
PDB header: transcription/dnaChain: E: PDB Molecule: p55-c-fos proto-oncogene protein;PDBTitle: two human c-fos:c-jun:dna complexes
27 c2w6aB_
not modelled
57.3
30
PDB header: signaling proteinChain: B: PDB Molecule: arf gtpase-activating protein git1;PDBTitle: x-ray structure of the dimeric git1 coiled-coil domain
28 c2r9vA_
not modelled
57.0
15
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase subunit alpha;PDBTitle: crystal structure of atp synthase subunit alpha (tm1612) from2 thermotoga maritima at 2.10 a resolution
29 c3c2qA_
not modelled
53.8
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized conserved protein;PDBTitle: crystal structure of conserved putative lor/sdh protein2 from methanococcus maripaludis s2
30 c2jsnA_
not modelled
53.6
25
PDB header: protein transportChain: A: PDB Molecule: trafficking protein particle complex subunit 4;PDBTitle: solution structure of the atypical pdz-like domain of2 synbindin
31 d2f1la1
not modelled
52.5
22
Fold: PRC-barrel domainSuperfamily: PRC-barrel domainFamily: RimM C-terminal domain-like32 c1go4F_
not modelled
50.3
44
PDB header: cell cycleChain: F: PDB Molecule: mad1 (mitotic arrest deficient)-like 1;PDBTitle: crystal structure of mad1-mad2 reveals a conserved mad22 binding motif in mad1 and cdc20.
33 c2ve7A_
not modelled
50.2
21
PDB header: cell cycleChain: A: PDB Molecule: kinetochore protein hec1, kinetochore protein spc25;PDBTitle: crystal structure of a bonsai version of the human ndc802 complex
34 d1f6ga_
not modelled
47.6
9
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels35 c1rb1B_
not modelled
45.6
26
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
36 c3k7zA_
not modelled
45.6
26
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
37 c3k7zB_
not modelled
45.6
26
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
38 c1rb1A_
not modelled
45.6
26
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
39 c1swiA_
not modelled
45.6
26
PDB header: leucine zipperChain: A: PDB Molecule: gcn4p1;PDBTitle: gcn4-leucine zipper core mutant as n16a complexed with2 benzene
40 c1rb6C_
not modelled
45.6
26
PDB header: dna binding proteinChain: C: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as2 n16a tetragonal form
41 d1nppa2
not modelled
45.6
21
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain42 d1uklc_
not modelled
44.5
21
Fold: HLH-likeSuperfamily: HLH, helix-loop-helix DNA-binding domainFamily: HLH, helix-loop-helix DNA-binding domain43 c1ij2C_
not modelled
44.4
26
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvtl coiled-coil trimer with threonine at the a(16)2 position
44 d1nz9a_
not modelled
43.3
23
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain45 c1ij3C_
not modelled
43.3
26
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
46 c1ij3B_
not modelled
43.3
26
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
47 c2ywxA_
not modelled
42.8
13
PDB header: lyaseChain: A: PDB Molecule: phosphoribosylaminoimidazole carboxylase catalytic subunit;PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from methanocaldococcus jannaschii
48 c1ztaA_
not modelled
42.2
26
PDB header: dna-binding motifChain: A: PDB Molecule: leucine zipper monomer;PDBTitle: the solution structure of a leucine-zipper motif peptide
49 c1ij2B_
not modelled
42.2
26
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvtl coiled-coil trimer with threonine at the a(16)2 position
50 c3a5tB_
not modelled
41.5
26
PDB header: transcription regulator/dnaChain: B: PDB Molecule: transcription factor mafg;PDBTitle: crystal structure of mafg-dna complex
51 c2zdiC_
not modelled
41.3
18
PDB header: chaperoneChain: C: PDB Molecule: prefoldin subunit alpha;PDBTitle: crystal structure of prefoldin from pyrococcus horikoshii2 ot3
52 c2kvqG_
not modelled
40.9
23
PDB header: transcriptionChain: G: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of nuse:nusg-ctd complex
53 c2jvvA_
not modelled
40.9
23
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of e. coli nusg carboxyterminal domain
54 c1gk7A_
not modelled
40.6
17
PDB header: vimentinChain: A: PDB Molecule: vimentin;PDBTitle: human vimentin coil 1a fragment (1a)
55 c2qe7C_
not modelled
40.0
23
PDB header: hydrolaseChain: C: PDB Molecule: atp synthase subunit alpha;PDBTitle: crystal structure of the f1-atpase from the thermoalkaliphilic2 bacterium bacillus sp. ta2.a1
56 c3q4fG_
not modelled
39.5
17
PDB header: dna binding protein/protein bindingChain: G: PDB Molecule: dna repair protein xrcc4;PDBTitle: crystal structure of xrcc4/xlf-cernunnos complex
57 c1ik9B_
not modelled
39.5
16
PDB header: gene regulation/ligaseChain: B: PDB Molecule: dna repair protein xrcc4;PDBTitle: crystal structure of a xrcc4-dna ligase iv complex
58 c2o7hF_
not modelled
38.1
23
PDB header: transcriptionChain: F: PDB Molecule: general control protein gcn4;PDBTitle: crystal structure of trimeric coiled coil gcn4 leucine zipper
59 c1ci6A_
not modelled
37.0
20
PDB header: transcriptionChain: A: PDB Molecule: transcription factor atf-4;PDBTitle: transcription factor atf4-c/ebp beta bzip heterodimer
60 d1am9a_
not modelled
36.6
21
Fold: HLH-likeSuperfamily: HLH, helix-loop-helix DNA-binding domainFamily: HLH, helix-loop-helix DNA-binding domain61 c2nn6I_
not modelled
35.5
16
PDB header: hydrolase/transferaseChain: I: PDB Molecule: 3'-5' exoribonuclease csl4 homolog;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
62 d1xi8a3
not modelled
35.3
17
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like63 c1fosF_
not modelled
35.1
14
PDB header: transcription/dnaChain: F: PDB Molecule: c-jun proto-oncogene protein;PDBTitle: two human c-fos:c-jun:dna complexes
64 c2gd7B_
not modelled
34.6
29
PDB header: transcriptionChain: B: PDB Molecule: hexim1 protein;PDBTitle: the structure of the cyclin t-binding domain of hexim12 reveals the molecular basis for regulation of3 transcription elongation
65 d2z1ca1
not modelled
33.1
25
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like66 c1w5kB_
not modelled
32.0
35
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: an anti-parallel four helix bundle
67 c1w5kC_
not modelled
32.0
35
PDB header: four helix bundleChain: C: PDB Molecule: general control protein gcn4;PDBTitle: an anti-parallel four helix bundle
68 c1w5kD_
not modelled
32.0
35
PDB header: four helix bundleChain: D: PDB Molecule: general control protein gcn4;PDBTitle: an anti-parallel four helix bundle
69 c1w5kA_
not modelled
32.0
35
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: an anti-parallel four helix bundle
70 c1unxA_
not modelled
31.0
29
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
71 c2j66A_
not modelled
31.0
20
PDB header: lyaseChain: A: PDB Molecule: btrk;PDBTitle: structural characterisation of btrk decarboxylase from2 butirosin biosynthesis
72 c2w5eB_
not modelled
30.6
19
PDB header: hydrolaseChain: B: PDB Molecule: putative serine protease;PDBTitle: structural and biochemical analysis of human pathogenic2 astrovirus serine protease at 2.0 angstrom resolution
73 c1p9iA_
not modelled
30.4
23
PDB header: unknown functionChain: A: PDB Molecule: cortexillin i/gcn4 hybrid peptide;PDBTitle: coiled-coil x-ray structure at 1.17 a resolution
74 c3ipkA_
not modelled
30.0
18
PDB header: cell adhesionChain: A: PDB Molecule: agi/ii;PDBTitle: crystal structure of a3vp1 of agi/ii of streptococcus mutans
75 d2nn6i1
not modelled
29.9
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like76 c2je6I_
not modelled
29.9
18
PDB header: hydrolaseChain: I: PDB Molecule: exosome complex rna-binding protein 1;PDBTitle: structure of a 9-subunit archaeal exosome
77 c1gclC_
not modelled
29.6
29
PDB header: leucine zipperChain: C: PDB Molecule: gcn4;PDBTitle: gcn4 leucine zipper core mutant p-li
78 c1gclB_
not modelled
29.6
29
PDB header: leucine zipperChain: B: PDB Molecule: gcn4;PDBTitle: gcn4 leucine zipper core mutant p-li
79 c1gclA_
not modelled
29.6
29
PDB header: leucine zipperChain: A: PDB Molecule: gcn4;PDBTitle: gcn4 leucine zipper core mutant p-li
80 c3bboW_
not modelled
29.5
25
PDB header: ribosomeChain: W: PDB Molecule: ribosomal protein l24;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
81 c1w5jD_
not modelled
29.3
35
PDB header: four helix bundleChain: D: PDB Molecule: general control protein gcn4;PDBTitle: an anti-parallel four helix bundle
82 c1w5jA_
not modelled
29.3
35
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: an anti-parallel four helix bundle
83 c1w5jB_
not modelled
29.3
35
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: an anti-parallel four helix bundle
84 c1w5jC_
not modelled
29.3
35
PDB header: four helix bundleChain: C: PDB Molecule: general control protein gcn4;PDBTitle: an anti-parallel four helix bundle
85 c2xv5A_
not modelled
29.1
24
PDB header: structural proteinChain: A: PDB Molecule: lamin-a/c;PDBTitle: human lamin a coil 2b fragment
86 c2rjzA_
not modelled
29.1
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pilo protein;PDBTitle: crystal structure of the type 4 fimbrial biogenesis protein pilo from2 pseudomonas aeruginosa
87 c3e98B_
not modelled
28.1
28
PDB header: unknown functionChain: B: PDB Molecule: gaf domain of unknown function;PDBTitle: crystal structure of a gaf domain containing protein that belongs to2 pfam duf484 family (pa5279) from pseudomonas aeruginosa at 2.43 a3 resolution
88 d1fx0b2
not modelled
28.1
19
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase89 c1kmhA_
not modelled
28.0
14
PDB header: hydrolaseChain: A: PDB Molecule: atpase alpha subunit;PDBTitle: crystal structure of spinach chloroplast f1-atpase2 complexed with tentoxin
90 c3e0eA_
not modelled
27.9
14
PDB header: replicationChain: A: PDB Molecule: replication protein a;PDBTitle: crystal structure of a domain of replication protein a from2 methanococcus maripaludis. northeast structural genomics3 targe mrr110b
91 c2dyiA_
not modelled
27.7
32
PDB header: ribosomeChain: A: PDB Molecule: probable 16s rrna-processing protein rimm;PDBTitle: crystal structure of 16s ribosomal rna processing protein rimm from2 thermus thermophilus hb8
92 c1w5iA_
not modelled
27.7
29
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: aba does not affect topology of pli.
93 c1uo2A_
not modelled
27.7
29
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
94 c1uo1B_
not modelled
27.7
29
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
95 c1unwB_
not modelled
27.6
29
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
96 c2ccfA_
not modelled
27.6
29
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel configuration of pli e20s
97 c1uo0A_
not modelled
27.6
29
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
98 c1uo0B_
not modelled
27.6
29
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
99 d1x87a_
not modelled
27.5
50
Fold: UrocanaseSuperfamily: UrocanaseFamily: Urocanase100 c1unxB_
not modelled
27.5
29
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
101 d1skye2
not modelled
27.4
29
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase102 d1mabb2
not modelled
27.3
24
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase103 c2w6fA_
not modelled
27.2
15
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase subunit alpha heart isoform,PDBTitle: low resolution structures of bovine mitochondrial f1-atpase2 during controlled dehydration: hydration state 2.
104 d2oa5a1
not modelled
27.0
17
Fold: BLRF2-likeSuperfamily: BLRF2-likeFamily: BLRF2-like105 d2jdid2
not modelled
26.9
24
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase106 c1w5iB_
not modelled
26.2
29
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: aba does not affect topology of pli.
107 c1uo2B_
not modelled
26.2
29
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
108 d1uwka_
not modelled
25.5
46
Fold: UrocanaseSuperfamily: UrocanaseFamily: Urocanase109 c3trhI_
not modelled
25.1
12
PDB header: lyaseChain: I: PDB Molecule: phosphoribosylaminoimidazole carboxylasePDBTitle: structure of a phosphoribosylaminoimidazole carboxylase catalytic2 subunit (pure) from coxiella burnetii
110 c3h9nA_
not modelled
24.5
25
PDB header: ribosomal proteinChain: A: PDB Molecule: ribosome maturation factor rimm;PDBTitle: crystal structure of the ribosome maturation factor rimm2 (hi0203) from h.influenzae. northeast structural genomics3 consortium target ir66.
111 d1j2za_
not modelled
24.5
9
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: UDP N-acetylglucosamine acyltransferase112 c2f1lA_
not modelled
23.6
18
PDB header: unknown functionChain: A: PDB Molecule: 16s rrna processing protein;PDBTitle: crystal structure of a putative 16s ribosomal rna processing protein2 rimm (pa3744) from pseudomonas aeruginosa at 2.46 a resolution
113 c1gd2G_
not modelled
23.4
23
PDB header: transcription/dnaChain: G: PDB Molecule: transcription factor pap1;PDBTitle: crystal structure of bzip transcription factor pap1 bound2 to dna
114 d2otma1
not modelled
23.3
26
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP115 c3l31B_
not modelled
23.3
38
PDB header: hydrolaseChain: B: PDB Molecule: probable manganase-dependent inorganicPDBTitle: crystal structure of the cbs and drtgg domains of the2 regulatory region of clostridium perfringens3 pyrophosphatase complexed with the inhibitor, amp
116 c2vpmB_
not modelled
23.2
25
PDB header: ligaseChain: B: PDB Molecule: trypanothione synthetase;PDBTitle: trypanothione synthetase
117 d1wzua1
not modelled
22.9
19
Fold: NadA-likeSuperfamily: NadA-likeFamily: NadA-like118 d1jjcb3
not modelled
22.9
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Myf domain119 c2p3eA_
not modelled
22.9
25
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal structure of aq1208 from aquifex aeolicus
120 d2jf2a1
not modelled
22.8
0
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: UDP N-acetylglucosamine acyltransferase