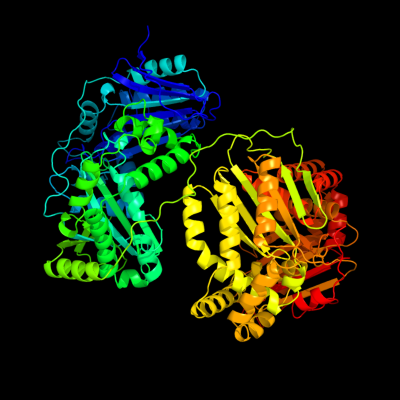
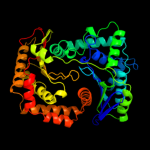
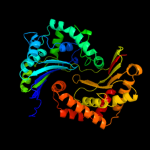
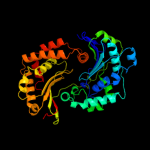
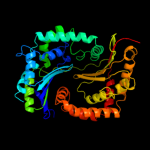
1 c1q2lA_
100.0
100
PDB header: hydrolaseChain: A: PDB Molecule: protease iii;PDBTitle: crystal structure of pitrilysin
2 c2jbuB_
100.0
27
PDB header: hydrolaseChain: B: PDB Molecule: insulin-degrading enzyme;PDBTitle: crystal structure of human insulin degrading enzyme2 complexed with co-purified peptides.
3 c2wk3A_
100.0
28
PDB header: hydrolaseChain: A: PDB Molecule: insulin degrading enzyme;PDBTitle: crystal structure of human insulin-degrading enzyme in2 complex with amyloid-beta (1-42)
4 c2fgeA_
100.0
13
PDB header: hydrolase, plant proteinChain: A: PDB Molecule: zinc metalloprotease (insulinase family);PDBTitle: crystal structure of presequence protease prep from2 arabidopsis thaliana
5 c1hr9D_
100.0
17
PDB header: hydrolaseChain: D: PDB Molecule: mitochondrial processing peptidase beta subunit;PDBTitle: yeast mitochondrial processing peptidase beta-e73q mutant2 complexed with malate dehydrogenase signal peptide
6 c1sqpA_
100.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: ubiquinol-cytochrome-c reductase complex core protein i,PDBTitle: crystal structure analysis of bovine bc1 with myxothiazol
7 c3eoqB_
100.0
16
PDB header: hydrolaseChain: B: PDB Molecule: putative zinc protease;PDBTitle: the crystal structure of putative zinc protease beta-2 subunit from thermus thermophilus hb8
8 c3amiB_
100.0
16
PDB header: hydrolaseChain: B: PDB Molecule: zinc peptidase;PDBTitle: the crystal structure of the m16b metallopeptidase subunit from2 sphingomonas sp. a1
9 c1nu1A_
100.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: ubiquinol-cytochrome c reductase complex core protein i,PDBTitle: crystal structure of mitochondrial cytochrome bc1 complexed with 2-2 nonyl-4-hydroxyquinoline n-oxide (nqno)
10 c3go9A_
100.0
14
PDB header: hydrolaseChain: A: PDB Molecule: insulinase family protease;PDBTitle: predicted insulinase family protease from yersinia pestis
11 c1hr6C_
100.0
12
PDB header: hydrolaseChain: C: PDB Molecule: mitochondrial processing peptidase alpha subunit;PDBTitle: yeast mitochondrial processing peptidase
12 c3hdiA_
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: processing protease;PDBTitle: crystal structure of bacillus halodurans metallo peptidase
13 c1l0lB_
100.0
11
PDB header: oxidoreductaseChain: B: PDB Molecule: ubiquinol-cytochrome c reductase complex core protein 2;PDBTitle: structure of bovine mitochondrial cytochrome bc1 complex with a bound2 fungicide famoxadone
14 c3cx5L_
100.0
15
PDB header: oxidoreductaseChain: L: PDB Molecule: cytochrome b-c1 complex subunit 1, mitochondrial;PDBTitle: structure of complex iii with bound cytochrome c in reduced2 state and definition of a minimal core interface for3 electron transfer.
15 c3gwbA_
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: peptidase m16 inactive domain family protein;PDBTitle: crystal structure of peptidase m16 inactive domain from pseudomonas2 fluorescens. northeast structural genomics target plr293l
16 c3amjB_
100.0
16
PDB header: hydrolaseChain: B: PDB Molecule: zinc peptidase inactive subunit;PDBTitle: the crystal structure of the heterodimer of m16b peptidase from2 sphingomonas sp. a1
17 c3d3yA_
100.0
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a conserved protein from enterococcus faecalis2 v583
18 d1q2la4
100.0
100
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like19 d2fgea4
100.0
22
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like20 d1ppja1
100.0
22
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like21 d1hr6b1
not modelled
100.0
24
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like22 d1hr6a1
not modelled
100.0
13
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like23 c3cxhM_
not modelled
100.0
14
PDB header: oxidoreductaseChain: M: PDB Molecule: cytochrome b-c1 complex subunit 2, mitochondrial;PDBTitle: structure of yeast complex iii with isoform-2 cytochrome c2 bound and definition of a minimal core interface for3 electron transfer.
24 d1bcca1
not modelled
100.0
22
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like25 d1q2la2
not modelled
100.0
100
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like26 d1ppjb1
not modelled
100.0
13
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like27 d1bccb1
not modelled
100.0
13
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like28 d3cx5a1
not modelled
100.0
15
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like29 d3cx5b1
not modelled
100.0
16
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like30 d1q2la3
not modelled
100.0
100
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like31 d1q2la1
not modelled
100.0
100
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like32 d1ppjb2
not modelled
99.8
12
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like33 d1bcca2
not modelled
99.8
7
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like34 d1hr6b2
not modelled
99.8
7
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like35 d1hr6a2
not modelled
99.8
11
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like36 d2fgea2
not modelled
99.8
9
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like37 c3ivlA_
not modelled
99.8
12
PDB header: hydrolaseChain: A: PDB Molecule: putative zinc protease;PDBTitle: the crystal structure of the inactive peptidase domain of a putative2 zinc protease from bordetella parapertussis to 2.2a
38 d1ppja2
not modelled
99.8
6
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like39 d1bccb2
not modelled
99.7
12
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like40 d3cx5a2
not modelled
99.7
12
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like41 d2fgea3
not modelled
99.6
12
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like42 d2fgea1
not modelled
99.2
10
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like43 c3ffvA_
not modelled
52.1
13
PDB header: protein bindingChain: A: PDB Molecule: protein syd;PDBTitle: crystal structure analysis of syd
44 d1j96a_
not modelled
37.5
31
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)45 d2rdea1
not modelled
22.0
18
Fold: Split barrel-likeSuperfamily: PilZ domain-likeFamily: PilZ domain46 c2rd9C_
not modelled
20.5
14
PDB header: hydrolaseChain: C: PDB Molecule: bh0186 protein;PDBTitle: crystal structure of a putative yfit-like metal-dependent hydrolase2 (bh0186) from bacillus halodurans c-125 at 2.30 a resolution
47 c3hrmA_
not modelled
18.2
26
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional regulator sarz;PDBTitle: crystal structure of staphylococcus aureus protein sarz in sulfenic2 acid form
48 d1jgsa_
not modelled
17.7
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators49 d1lb3a_
not modelled
13.8
16
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin50 c3ostA_
not modelled
12.9
10
PDB header: lipid binding proteinChain: A: PDB Molecule: serine/threonine-protein kinase kcc4;PDBTitle: structure of the kinase associated-1 (ka1) from kcc4p
51 c2kfvA_
not modelled
12.9
25
PDB header: isomeraseChain: A: PDB Molecule: fk506-binding protein 3;PDBTitle: structure of the amino-terminal domain of human fk506-2 binding protein 3 / northeast structural genomics3 consortium target ht99a
52 c2zkr9_
not modelled
12.1
21
PDB header: ribosomal protein/rnaChain: 9: PDB Molecule: 60s ribosomal protein l32;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
53 d1r03a_
not modelled
11.9
18
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin54 d1xl7a1
not modelled
10.9
15
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase55 c3l7vA_
not modelled
10.1
13
PDB header: transcriptionChain: A: PDB Molecule: putative uncharacterized protein smu.1377c;PDBTitle: crystal structure of a hypothetical protein smu.1377c from2 streptococcus mutans ua159
56 d3broa1
not modelled
9.7
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators57 d1u1qa_
not modelled
9.7
4
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD58 c1xgwA_
not modelled
9.6
14
PDB header: endocytosisChain: A: PDB Molecule: epsin 4;PDBTitle: the crystal structure of human enthoprotin n-terminal domain
59 c3g3zA_
not modelled
9.4
19
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, marr family;PDBTitle: the structure of nmb1585, a marr family regulator from neisseria2 meningitidis
60 d1qkia1
not modelled
9.3
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain61 c3llkA_
not modelled
8.9
16
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfhydryl oxidase 1;PDBTitle: sulfhydryl oxidase fragment of human qsox1
62 c2pbrB_
not modelled
8.7
12
PDB header: transferaseChain: B: PDB Molecule: thymidylate kinase;PDBTitle: crystal structure of thymidylate kinase (aq_969) from aquifex aeolicus2 vf5
63 c2qm6C_
not modelled
8.7
17
PDB header: transferaseChain: C: PDB Molecule: gamma-glutamyltranspeptidase;PDBTitle: crystal structure of helicobacter pylori gamma-glutamyltranspeptidase2 in complex with glutamate
64 d1p4xa1
not modelled
8.5
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators65 c2uxsA_
not modelled
8.4
14
PDB header: hydrolaseChain: A: PDB Molecule: inorganic pyrophosphatase;PDBTitle: 2.7a crystal structure of inorganic pyrophosphatase (rv3628)2 from mycobacterium tuberculosis at ph 7.5
66 c3onlB_
not modelled
8.3
15
PDB header: protein transportChain: B: PDB Molecule: epsin-3;PDBTitle: yeast ent3_enth-vti1p_habc complex structure
67 d1y0ga_
not modelled
8.1
19
Fold: Streptavidin-likeSuperfamily: YceI-likeFamily: YceI-like68 d1r62a_
not modelled
8.1
19
Fold: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseSuperfamily: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinaseFamily: Histidine kinase69 d1mfra_
not modelled
8.0
15
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin70 d1eyha_
not modelled
8.0
14
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: ENTH domain71 d1z6om1
not modelled
8.0
24
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin72 c3iz5h_
not modelled
7.8
14
PDB header: ribosomeChain: H: PDB Molecule: 60s ribosomal protein l7a (l7ae);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
73 d2fbha1
not modelled
7.7
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators74 d3c7ba2
not modelled
7.7
11
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: DsrA/DsrB N-terminal-domain-like75 c4a1cX_
not modelled
7.7
18
PDB header: ribosomeChain: X: PDB Molecule: 60s ribosomal protein l32;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
76 d1udea_
not modelled
7.6
9
Fold: OB-foldSuperfamily: Inorganic pyrophosphataseFamily: Inorganic pyrophosphatase77 c3nnhA_
not modelled
7.5
15
PDB header: rna binding protein/rnaChain: A: PDB Molecule: cugbp elav-like family member 1;PDBTitle: crystal structure of the cugbp1 rrm1 with guuguuuuguuu rna
78 c2adbA_
not modelled
7.5
14
PDB header: rna binding protein/rnaChain: A: PDB Molecule: polypyrimidine tract-binding protein 1;PDBTitle: solution structure of polypyrimidine tract binding protein2 rbd2 complexed with cucucu rna
79 d1l3ka1
not modelled
7.4
4
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD80 c2hvzA_
not modelled
7.4
14
PDB header: rna binding proteinChain: A: PDB Molecule: splicing factor, arginine/serine-rich 7;PDBTitle: solution structure of the rrm domain of sr rich factor 9g8
81 d2ceia1
not modelled
7.4
19
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin82 d1zavz1
not modelled
7.3
32
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain83 c1zavZ_
not modelled
7.3
32
PDB header: structural proteinChain: Z: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
84 c1zawZ_
not modelled
7.3
32
PDB header: structural proteinChain: Z: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
85 c1zawX_
not modelled
7.2
30
PDB header: structural proteinChain: X: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
86 c1zawY_
not modelled
7.2
30
PDB header: structural proteinChain: Y: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
87 d2za7a1
not modelled
7.2
16
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ferritin88 c3ns5B_
not modelled
7.2
26
PDB header: translationChain: B: PDB Molecule: eukaryotic translation initiation factor 3 subunit b;PDBTitle: crystal structure of the rna recognition motif of yeast eif3b residues2 76-161
89 c1xl8B_
not modelled
7.1
14
PDB header: transferaseChain: B: PDB Molecule: peroxisomal carnitine o-octanoyltransferase;PDBTitle: crystal structure of mouse carnitine octanoyltransferase in2 complex with octanoylcarnitine
90 d1whxa_
not modelled
7.1
29
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD91 c2x3dC_
not modelled
7.0
14
PDB header: unknown functionChain: C: PDB Molecule: sso6206;PDBTitle: crystal structure of sso6206 from sulfolobus solfataricus p2
92 c3lv8A_
not modelled
7.0
2
PDB header: transferaseChain: A: PDB Molecule: thymidylate kinase;PDBTitle: 1.8 angstrom resolution crystal structure of a thymidylate kinase2 (tmk) from vibrio cholerae o1 biovar eltor str. n16961 in complex3 with tmp, thymidine-5'-diphosphate and adp
93 c2qy7A_
not modelled
7.0
12
PDB header: protein bindingChain: A: PDB Molecule: clathrin interactor 1;PDBTitle: crystal structure of human epsinr enth domain
94 d1lnwa_
not modelled
7.0
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators95 c2rs2A_
not modelled
6.9
9
PDB header: rna binding protein/rnaChain: A: PDB Molecule: rna-binding protein musashi homolog 1;PDBTitle: 1h, 13c, and 15n chemical shift assignments for musashi1 rbd1:r(guagu)2 complex
96 c2fa5B_
not modelled
6.8
13
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator marr/emrr family;PDBTitle: the crystal structure of an unliganded multiple antibiotic-2 resistance repressor (marr) from xanthomonas campestris
97 c1ygaA_
not modelled
6.7
10
PDB header: isomeraseChain: A: PDB Molecule: hypothetical 37.9 kda protein in bio3-hxt17PDBTitle: crystal structure of saccharomyces cerevisiae yn9a protein,2 new york structural genomics consortium
98 d1hsja1
not modelled
6.6
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators99 c1qapA_
not modelled
6.6
16
PDB header: glycosyltransferaseChain: A: PDB Molecule: quinolinic acid phosphoribosyltransferase;PDBTitle: quinolinic acid phosphoribosyltransferase with bound2 quinolinic acid