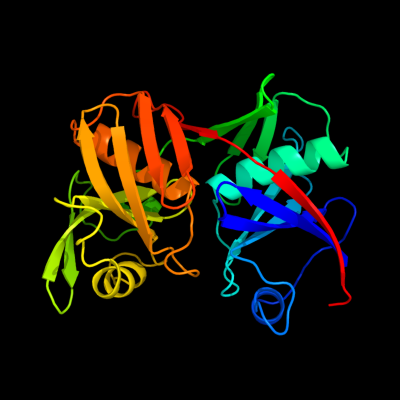
1 c2gkjA_
100.0
77
PDB header: isomeraseChain: A: PDB Molecule: diaminopimelate epimerase;PDBTitle: crystal structure of diaminopimelate epimerase in complex2 with an irreversible inhibitor dl-azidap
2 c3ekmE_
100.0
41
PDB header: isomeraseChain: E: PDB Molecule: diaminopimelate epimerase, chloroplastic;PDBTitle: crystal structure of diaminopimelate epimerase form2 arabidopsis thaliana in complex with irreversible inhibitor3 dl-azidap
3 c3fveA_
100.0
31
PDB header: isomeraseChain: A: PDB Molecule: diaminopimelate epimerase;PDBTitle: crystal structure of diaminopimelate epimerase mycobacterium2 tuberculosis dapf
4 c2otnB_
100.0
33
PDB header: isomeraseChain: B: PDB Molecule: diaminopimelate epimerase;PDBTitle: crystal structure of the catalytically active form of diaminopimelate2 epimerase from bacillus anthracis
5 c3ednB_
100.0
20
PDB header: biosynthetic proteinChain: B: PDB Molecule: phenazine biosynthesis protein, phzf family;PDBTitle: crystal structure of the bacillus anthracis phenazine2 biosynthesis protein, phzf family
6 c1qy9B_
100.0
17
PDB header: unknown functionChain: B: PDB Molecule: hypothetical protein ydde;PDBTitle: crystal structure of e. coli se-met protein ydde
7 c1ym5A_
100.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: hypothetical 32.6 kda protein in dap2-slt2PDBTitle: crystal structure of yhi9, the yeast member of the2 phenazine biosynthesis phzf enzyme superfamily.
8 d1s7ja_
100.0
14
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like9 c1u1wA_
100.0
20
PDB header: isomerase, lyaseChain: A: PDB Molecule: phenazine biosynthesis protein phzf;PDBTitle: structure and function of phenazine-biosynthesis protein phzf from2 pseudomonas fluorescens 2-79
10 c1u0kA_
100.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: gene product pa4716;PDBTitle: the structure of a predicted epimerase pa4716 from pseudomonas2 aeruginosa
11 c2azpA_
100.0
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pa1268;PDBTitle: crystal structure of pa1268 solved by sulfur sad
12 d2gkea1
100.0
81
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: Diaminopimelate epimerase13 c1w62B_
100.0
15
PDB header: racemaseChain: B: PDB Molecule: b-cell mitogen;PDBTitle: proline racemase in complex with one molecule of pyrrole-2-2 carboxylic acid (hemi form)
14 d1tm0a_
100.0
17
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: Proline racemase15 d2gkea2
100.0
75
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: Diaminopimelate epimerase16 d1qy9a1
99.9
21
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like17 d1xuba1
99.9
17
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like18 d1xuba2
99.9
20
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like19 d1u0ka1
99.9
18
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like20 d1qy9a2
99.8
15
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like21 d1u0ka2
not modelled
99.7
20
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like22 c3g7kD_
not modelled
98.6
16
PDB header: isomeraseChain: D: PDB Molecule: 3-methylitaconate isomerase;PDBTitle: crystal structure of methylitaconate-delta-isomerase
23 c2pw0A_
not modelled
98.4
20
PDB header: unknown functionChain: A: PDB Molecule: prpf methylaconitate isomerase;PDBTitle: crystal structure of trans-aconitate bound to methylaconitate2 isomerase prpf from shewanella oneidensis
24 d2h9fa2
not modelled
96.6
21
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PA0793-like25 d2h9fa1
not modelled
61.8
15
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PA0793-like26 c3g12A_
not modelled
39.2
11
PDB header: lyaseChain: A: PDB Molecule: putative lactoylglutathione lyase;PDBTitle: crystal structure of a putative lactoylglutathione lyase2 from bdellovibrio bacteriovorus
27 c3ldgA_
not modelled
36.9
19
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein smu.472;PDBTitle: crystal structure of smu.472, a putative methyltransferase complexed2 with sah
28 c3sk1C_
not modelled
31.5
24
PDB header: griseoluteate-binding proteinChain: C: PDB Molecule: ehpr;PDBTitle: crystal structure of phenazine resistance protein ehpr from2 enterobacter agglomerans (erwinia herbicola, pantoea agglomerans)3 eh1087, apo form
29 c2l3bA_
not modelled
30.0
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein found in conjugate transposon;PDBTitle: solution nmr structure of the bt_0084 lipoprotein from bacteroides2 thetaiotaomicron, northeast structural genomics consortium target3 btr376
30 d1o9ga_
not modelled
29.6
20
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: rRNA methyltransferase AviRa31 c2l7qA_
not modelled
27.9
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein found in conjugate transposon;PDBTitle: solution nmr structure of conjugate transposon protein bvu_1572(27-2 141) from bacteroides vulgatus, northeast structural genomics3 consortium target bvr155
32 c3k0bA_
not modelled
26.0
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted n6-adenine-specific dna methylase;PDBTitle: crystal structure of a predicted n6-adenine-specific dna methylase2 from listeria monocytogenes str. 4b f2365
33 c1g38A_
not modelled
22.2
38
PDB header: transferase/dnaChain: A: PDB Molecule: modification methylase taqi;PDBTitle: adenine-specific methyltransferase m. taq i/dna complex
34 d2f8la1
not modelled
21.0
14
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: N-6 DNA Methylase-like35 d2ar0a1
not modelled
19.8
13
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: N-6 DNA Methylase-like36 d1c4qa_
not modelled
19.6
15
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits37 c3lkdB_
not modelled
19.0
38
PDB header: transferaseChain: B: PDB Molecule: type i restriction-modification systemPDBTitle: crystal structure of the type i restriction-modification2 system methyltransferase subunit from streptococcus3 thermophilus, northeast structural genomics consortium4 target sur80
38 c3lduA_
not modelled
17.8
14
PDB header: transferaseChain: A: PDB Molecule: putative methylase;PDBTitle: the crystal structure of a possible methylase from2 clostridium difficile 630.
39 d2ih2a1
not modelled
17.7
18
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: DNA methylase TaqI, N-terminal domain40 c2kjzA_
not modelled
17.0
10
PDB header: unknown functionChain: A: PDB Molecule: atc0852;PDBTitle: solution nmr structure of protein atc0852 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att2.
41 d1sr8a_
not modelled
16.4
19
Fold: CbiD-likeSuperfamily: CbiD-likeFamily: CbiD-like42 c1aqjB_
not modelled
15.6
21
PDB header: methyltransferaseChain: B: PDB Molecule: adenine-n6-dna-methyltransferase taqi;PDBTitle: structure of adenine-n6-dna-methyltransferase taqi
43 d1r4pb_
not modelled
15.4
17
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits44 c3owvA_
not modelled
14.9
16
PDB header: hydrolaseChain: A: PDB Molecule: dna-entry nuclease;PDBTitle: structural insights into catalytic and substrate binding mechanisms of2 the strategic enda nuclease from streptococcus pneumoniae
45 c2waqG_
not modelled
14.6
25
PDB header: transcriptionChain: G: PDB Molecule: dna-directed rna polymerase rpo8 subunit;PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
46 d2okca1
not modelled
13.9
26
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: N-6 DNA Methylase-like47 c3fd5B_
not modelled
13.8
18
PDB header: transferaseChain: B: PDB Molecule: selenide, water dikinase 1;PDBTitle: crystal structure of human selenophosphate synthetase 12 complex with ampcp
48 c3khkA_
not modelled
13.6
22
PDB header: dna binding proteinChain: A: PDB Molecule: type i restriction-modification systemPDBTitle: crystal structure of type-i restriction-modification system2 methylation subunit (mm_0429) from methanosarchina mazei.
49 d1h6za2
not modelled
13.1
18
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain50 c3hvzB_
not modelled
12.9
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the tgs domain of the clolep_03100 protein from2 clostridium leptum, northeast structural genomics consortium target3 qlr13a
51 c2xh3B_
not modelled
12.7
16
PDB header: hydrolaseChain: B: PDB Molecule: spd1 nuclease;PDBTitle: extracellular nuclease
52 c2kmmA_
not modelled
11.9
13
PDB header: hydrolaseChain: A: PDB Molecule: guanosine-3',5'-bis(diphosphate) 3'-PDBTitle: solution nmr structure of the tgs domain of pg1808 from2 porphyromonas gingivalis. northeast structural genomics3 consortium target pgr122a (418-481)
53 d1n2aa2
not modelled
11.3
11
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione S-transferase (GST), N-terminal domain54 c2xznC_
not modelled
10.7
12
PDB header: ribosomeChain: C: PDB Molecule: kh domain containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
55 c3p9nA_
not modelled
9.7
33
PDB header: transferaseChain: A: PDB Molecule: possible methyltransferase (methylase);PDBTitle: rv2966c of m. tuberculosis is a rsmd-like methyltransferase
56 c1wqaB_
not modelled
9.5
14
PDB header: isomeraseChain: B: PDB Molecule: phospho-sugar mutase;PDBTitle: crystal structure of pyrococcus horikoshii2 phosphomannomutase/phosphoglucomutase complexed with mg2+
57 d1twua_
not modelled
9.1
16
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Hypothetical protein YycE58 c3ghjA_
not modelled
9.0
4
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative integron gene cassette protein;PDBTitle: crystal structure from the mobile metagenome of halifax2 harbour sewage outfall: integron cassette protein hfx_cass4
59 c2ph0A_
not modelled
9.0
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the q6d2t7_erwct protein from erwinia2 carotovora. nesg target ewr41.
60 d2hqva1
not modelled
8.9
13
Fold: Heme iron utilization protein-likeSuperfamily: Heme iron utilization protein-likeFamily: ChuX-like61 c3r6aB_
not modelled
8.8
12
PDB header: isomerase, lyaseChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein (hypothetical protein2 mm_3218) from methanosarcina mazei.
62 c2zifB_
not modelled
8.6
21
PDB header: transferaseChain: B: PDB Molecule: putative modification methylase;PDBTitle: crystal structure of ttha0409, putative dna modification2 methylase from thermus thermophilus hb8- complexed with s-3 adenosyl-l-methionine
63 d1g60a_
not modelled
8.5
46
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Type II DNA methylase64 c2oviA_
not modelled
7.9
17
PDB header: ligand binding protein, metal transportChain: A: PDB Molecule: hypothetical protein chux;PDBTitle: structure of the heme binding protein chux
65 d1p3da1
not modelled
7.8
18
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain66 d2bosa_
not modelled
7.7
17
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits67 d1booa_
not modelled
7.5
31
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Type II DNA methylase68 c2f7lA_
not modelled
7.5
18
PDB header: isomeraseChain: A: PDB Molecule: 455aa long hypothetical phospho-sugar mutase;PDBTitle: crystal structure of sulfolobus tokodaii2 phosphomannomutase/phosphoglucomutase
69 c3m05A_
not modelled
7.4
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pepe_1480;PDBTitle: the crystal structure of a functionally unknown protein2 pepe_1480 from pediococcus pentosaceus atcc 25745
70 c3s1sA_
not modelled
7.3
20
PDB header: hydrolase, transferaseChain: A: PDB Molecule: restriction endonuclease bpusi;PDBTitle: characterization and crystal structure of the type iig restriction2 endonuclease bpusi
71 d1wy7a1
not modelled
6.4
20
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Ta1320-like72 d1m7va_
not modelled
6.3
18
Fold: Nitric oxide (NO) synthase oxygenase domainSuperfamily: Nitric oxide (NO) synthase oxygenase domainFamily: Nitric oxide (NO) synthase oxygenase domain73 d1ne2a_
not modelled
6.3
20
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Ta1320-like74 c2fkdK_
not modelled
6.2
30
PDB header: transcription regulatorChain: K: PDB Molecule: repressor protein ci;PDBTitle: crystal structure of the c-terminal domain of bacteriophage2 186 repressor
75 d2hiya1
not modelled
6.2
18
Fold: SP0830-likeSuperfamily: SP0830-likeFamily: SP0830-like76 c3gm5A_
not modelled
6.2
15
PDB header: isomeraseChain: A: PDB Molecule: lactoylglutathione lyase and related lyases;PDBTitle: crystal structure of a putative methylmalonyl-coenzyme a2 epimerase from thermoanaerobacter tengcongensis at 2.0 a3 resolution
77 c2h8bB_
not modelled
6.1
18
PDB header: hormone/growth factorChain: B: PDB Molecule: insulin-like 3;PDBTitle: solution structure of insl3
78 d1j3aa_
not modelled
6.1
10
Fold: Ribosomal protein L13Superfamily: Ribosomal protein L13Family: Ribosomal protein L1379 d1nhya2
not modelled
6.1
15
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione S-transferase (GST), N-terminal domain80 c1okgA_
not modelled
6.0
20
PDB header: transferaseChain: A: PDB Molecule: possible 3-mercaptopyruvate sulfurtransferase;PDBTitle: 3-mercaptopyruvate sulfurtransferase from leishmania major
81 c1nw6A_
not modelled
6.0
43
PDB header: transferaseChain: A: PDB Molecule: modification methylase rsri;PDBTitle: structure of the beta class n6-adenine dna methyltransferase rsri2 bound to sinefungin
82 d2esra1
not modelled
5.9
40
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: YhhF-like83 c1c4gB_
not modelled
5.6
12
PDB header: transferaseChain: B: PDB Molecule: protein (alpha-d-glucose 1-phosphatePDBTitle: phosphoglucomutase vanadate based transition state analog2 complex
84 d1hksa_
not modelled
5.5
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Heat-shock transcription factor85 d1xrka_
not modelled
5.5
7
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Antibiotic resistance proteins86 d1autl1
not modelled
5.4
43
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: EGF/LamininFamily: EGF-type module87 d1go4a_
not modelled
5.4
18
Fold: The spindle assembly checkpoint protein mad2Superfamily: The spindle assembly checkpoint protein mad2Family: The spindle assembly checkpoint protein mad288 c2f40A_
not modelled
5.3
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pf1455;PDBTitle: structure of a novel protein from backbone-centered nmr data and nmr-2 assisted structure prediction
89 d1xo8a_
not modelled
5.2
24
Fold: Immunoglobulin-like beta-sandwichSuperfamily: LEA14-likeFamily: LEA14-like