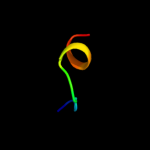| 1 | d2g7ja1
|
|
 |
100.0 |
88 |
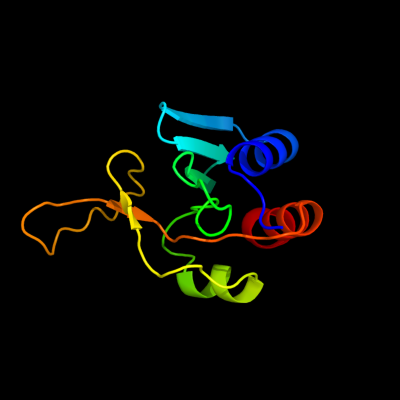
Fold:Secretion chaperone-like
Superfamily:YgaC/TfoX-N like
Family:YgaC-like |
| 2 | d1whza_
|
|
 |
38.4 |
32 |
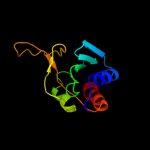
Fold:dsRBD-like
Superfamily:YcfA/nrd intein domain
Family:YcfA-like |
| 3 | d2odgc1
|
|
 |
18.6 |
83 |
Fold:LEM/SAP HeH motif
Superfamily:LEM domain
Family:LEM domain |
| 4 | d2h7aa1
|
|
 |
14.0 |
26 |
Fold:YcgL-like
Superfamily:YcgL-like
Family:YcgL-like |
| 5 | c3c2vA_
|
|
 |
13.5 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of the quinolinate phosphoribosyl2 transferase (bna6) from saccharomyces cerevisiae complexed3 with prpp and the inhibitor phthalate
|
| 6 | d2hqya1
|
|
 |
12.3 |
54 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:FemXAB nonribosomal peptidyltransferases |
| 7 | c3h0gN_
|
|
 |
11.7 |
23 |
PDB header:transcription
Chain: N: PDB Molecule:dna-directed rna polymerase ii subunit rpb2;
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
|
| 8 | d1twfb_
|
|
 |
10.5 |
24 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta |
| 9 | d1q8la_
|
|
 |
10.3 |
28 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 10 | d1smyc_
|
|
 |
9.8 |
36 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta |
| 11 | c3deoA_
|
|
 |
9.3 |
35 |
PDB header:protein transport, membrane protein
Chain: A: PDB Molecule:signal recognition particle 43 kda protein;
PDBTitle: structural basis for specific substrate recognition by the2 chloroplast signal recognition particle protein cpsrp43
|
| 12 | d1zyla1
|
|
 |
9.0 |
29 |
Fold:Protein kinase-like (PK-like)
Superfamily:Protein kinase-like (PK-like)
Family:APH phosphotransferases |
| 13 | c2pmzB_
|
|
 |
8.8 |
33 |
PDB header:translation, transferase
Chain: B: PDB Molecule:dna-directed rna polymerase subunit b;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
| 14 | c3dxsX_
|
|
 |
8.3 |
14 |
PDB header:hydrolase
Chain: X: PDB Molecule:copper-transporting atpase ran1;
PDBTitle: crystal structure of a copper binding domain from hma7, a p-2 type atpase
|
| 15 | d2gzsa1
|
|
 |
7.9 |
23 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:IroE-like |
| 16 | d1ynjc1
|
|
 |
6.7 |
38 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta |
| 17 | c2yh5A_
|
|
 |
6.6 |
40 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:dapx protein;
PDBTitle: structure of the c-terminal domain of bamc
|
| 18 | c3ktwA_
|
|
 |
6.4 |
25 |
PDB header:rna/rna binding protein
Chain: A: PDB Molecule:signal recognition particle 19 kda protein;
PDBTitle: crystal structure of the srp19/s-domain srp rna complex of sulfolobus2 solfataricus
|
| 19 | d1j5ja_
|
|
 |
6.2 |
58 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Scorpion toxin-like
Family:Short-chain scorpion toxins |
| 20 | c3pfhD_
|
|
 |
6.2 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:n-methyltransferase;
PDBTitle: x-ray crystal structure the n,n-dimethyltransferase tylm1 from2 streptomyces fradiae in complex with sah and dtdp-quip3n
|
| 21 | c2rogA_ |
|
not modelled |
6.1 |
22 |
PDB header:metal binding protein
Chain: A: PDB Molecule:heavy metal binding protein;
PDBTitle: solution structure of thermus thermophilus hb8 ttha17182 protein in living e. coli cells
|
| 22 | d1yrea1 |
|
not modelled |
6.0 |
17 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
| 23 | c1gxsC_ |
|
not modelled |
6.0 |
35 |
PDB header:lyase
Chain: C: PDB Molecule:p-(s)-hydroxymandelonitrile lyase chain a;
PDBTitle: crystal structure of hydroxynitrile lyase from sorghum2 bicolor in complex with inhibitor benzoic acid: a novel3 cyanogenic enzyme
|
| 24 | c3eywA_ |
|
not modelled |
5.9 |
25 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
| 25 | c3iydC_ |
|
not modelled |
5.7 |
33 |
PDB header:transcription/dna
Chain: C: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: three-dimensional em structure of an intact activator-dependent2 transcription initiation complex
|
| 26 | c2gcfA_ |
|
not modelled |
5.7 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:cation-transporting atpase pacs;
PDBTitle: solution structure of the n-terminal domain of the coppper(i) atpase2 pacs in its apo form
|
| 27 | d1civa2 |
|
not modelled |
5.7 |
26 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |